Linking GEOS-Chem to CMAQ
This page contains information about how to link the GEOS-Chem with the CMAQ regional air quality model.
Overview
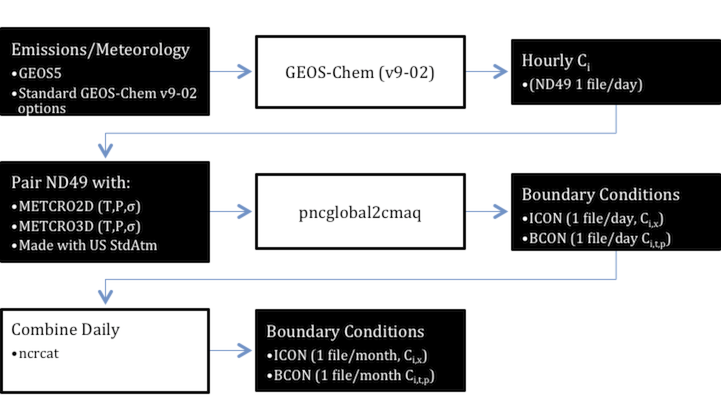
Global models are increasingly used as time-variable boundary conditions to regional models. Previously, representative concentrations (from models or observations) were used to construct time-invariant boundaries. This prevents important variability in the contribution of grid-internal and grid-external concentrations. By linking to global models, regional models conceptually improve the grid-external sources and provide their own estimates of grid-internal sources. To connect regional and global models, there are conceptually 4 steps:
- Horizontal Interpolation - Since most regional models use a curvilinear grid, this is the simplest step.
- Vertical Interpolation - Many regional models use a pure sigma-pressure coordinate, which is different from GEOS-Chem's hybrid eta-sigma.
- Tracer translation - GEOS-Chem uses explicit and lumped chemical species (gas and particle), which require one-to-one, many-to-one, and one-to-many mapping to regional model tracers.
- Unit conversions - Translated tracers must be converted to regional model units to ensure compatibility.
- Meta Data - Regional model meta-data must be provided to ensure compatibility.
Any utility that accomplishes these steps can be used.
pncglobal2cmaq
pncglobal2cmaq is part of the PseudoNetCDF software package (http://github.com/barronh/pseudonetcdf).
- Horizontal Interpolation - nearest neighbor with automatic conversion of CMAQ coordinates
- Pressure Interpolation - pressure vertical interpolation (recommended); pure eta is possible
- Tracer translation - Gas-phase translations for CB05 and SAPRC07 are provided; Aerosol translations to CMAQ AE5 and AE6 are provided; custom translations are also easy
- Unit conversion - dynamically converts units
- Meta Data - adds CMAQ meta-data to outputs automatically.
Instructions
A typical process has 4 steps and 2 pre-requisites. The pre-requisites are at least one CMAQ 3-D meteorology input for the inner domain (METCRO3D; only required for initial conditions) and one for the boundary (METBDY3D).
Run GEOS-Chem with ND49 enabled for the domain of interest
Create a copy of the chemical translation
pncglobal2cmaq.py --template=cb05tucl_ae6_aq /path/to/anyfile > mappings.json
- "--template" can be set to cb05tucl_ae5_aq, cb05tucl_ae6_aq, saprc07_ae5_aq, saprc07_ae6_aq
- mappings.json can be modified as desired.
Process one or more days ND49 file
pncglobal2cmaq.py nd49/ts20100227.bpch --METBDY3D met/METBDY3D_20100227 --METCRO3D met/METCRO3D_20100227 --mapping=mappings.json --outifolder=icon/ --outbfolder=bcon/
- Folders nd49, met, bcon, and icon are not required -- adjust paths to be consistent with your setup.
- "mappings.json" is the path that you created in the last step.
- "--METCRO3D" is only necessary to create an initial condition file and can be omitted for any
- "--METBDY3D" provides temperature and pressure for vertical interpolation and unit conversion.
Combine for CMAQ Simulation
CMAQ boundary files must have 25 instantaneous times per CMAQ simulation day. GEOS-Chem ND49 files typically have 24 times. Combine multiple days of output to ensure coverage (too much is okay) of all instantaneous times in CMAQ simulation. It is typical to use monthly BCON files for long simulations.
Combine Using PseudoNetCDF
pncgen --stack=TSTEP bcon/ts20100227.bpch.BCON.nc bcon/ts20100228.bpch.BCON.nc ... bcon/ts2010041.bpch.BCON.nc bcon/ts201003.BCON.nc
Combine Using NCO
ncrcat bcon/ts20100227.bpch.BCON.nc bcon/ts20100228.bpch.BCON.nc ... bcon/ts2010041.bpch.BCON.nc bcon/ts201003.BCON.nc
Output Visualization
Output files can be visualized using the PseudoNetCDF pncboundaries.py script. Below is a comparison of the CMAQ v5.0.2 standard BCON from "profiles" compared to the GEOS-Chem derived BCON. Note in GEOS-Chem, increasing ozone with altitude is seen as expected. PROFILE-based BCON has much lower ozone at high altitude because the data was collated before CMAQ top pressures were typically as high as they are now.
[[[File:Pncglobal2cmaq_comparison.png]]]
GEOSCHEM4CMAQ
Brief description
Tao Zeng wrote:
- The code to convert GEOS-Chem simulations as the boundary and initial conditions for CMAQ CB05 is ready. I have tested it successfully on GEOS-Chem v08-01-04 using pgf90 compiler. Some highlights are:
- (1) 2 output options:
- (a) in CMAQ domain after regridding. The outputs are CMAQ ready BC/IC files with NetCDF format. The MCIP met file (GRIDCRO3D and GRIDCRO2D) and default CMAQ IC/BC are required. IOAPI and NetCDF libs are also needed.
- (b) in G-C subdomain without regridding. It is in binary format.
- (2) Conversion of GOS-Chem species to CB05 tracers
- Tracer mapping table are designed to convert GEOS-Chem species to CMAQ tracers. Rob Pinder from EPA helped me to review it.
- CSPEC array is used to get direct NO and NO2 concentrations. Some other species in CSPEC but not in STT are also included. Several inactive species (ACTA, EOH, HCOOH, and MOH) are turned on into active species to get better mapping of CB05 tracers. This resulting differences are <1% in average over the domain after 1 month integration. And the max difference is ~2% for most tracers. But for NH3 and NIT, the max differences are up to 50% when they are at very low level.
- (3) To avoid excess downward transport from stratosphere in CMAQ, tracer concentrations at the tropopause in GEOS-Chem are used to for the tropopause and the layers above.
- Any suggestions are welcome. The whole package includes a new fortran module file and some lines adding into 3 files. If anybody is interested, please let me know.
--Bob Y. 10:19, 10 March 2010 (EST)
Authors and collaborators
- Tao Zeng (Georgia EPD)
User groups
| User Group | Personnel | Projects |
|---|---|---|
| Georgia EPD | Tao Zeng | ... |
| EPA | Havala Pye | Boundary conditions with a focus on aerosols |
| Add yours here... | ... | ... |
Source code
Tao Zeng has provided the GEOS-Chem to CMAQ source code linkage. It is available for download from:
ftp ftp.as.harvard.edu get pub/geos-chem/downloads/geoschem4cmaq.20100316.tgz
To extract the archive, type:
tar xvzf geoschem4cmaq.20100316.tgz
You will find a README.txt file in the archive with more information.
--Bob Y. 15:27, 16 March 2010 (EDT)
Validation
References
Known issues
SOA1, SOA2, SOA3, and SOA4 in G-C are outputed but are not converted to CB05 tracers. In CMAQ v4.7, 19 SOA species are defined. The CMAQ release notes do not fully explain them. So I will update this package when possible.
CMAQv5.0
Mapping for CMAQv5.0 with AERO6 including SOA --havala 16:14, 8 September 2011 (EDT)