Leaf area indices in GEOS-Chem
On this page we provide an overview of how leaf area index (LAI) data is used in GEOS-Chem. Leaf area index is defined as the total leaf surface area divided by the total surface area of a grid box (typical units are: cm2 leaf cm-2 GEOS-Chem grid box).
Also be sure to see our MODIS leaf area indices page, which describes the leaf area indices derived from the MODIS satellite instrument.
Contents
Overview
Prior to the official release of GEOS-Chem v9-01-03, GEOS-Chem used leaf area index data in the following manner:
LAI used by dry deposition and soil NOx emissions
As of this writing (April 2012), GEOS-Chem currently uses leaf area index (LAI) data from separate sources. The dry deposition and soil NOx emissions modules utilize the data stored in ASCII files named lai*.global. These files (one per month) are created (using offline IDL routines) for each specific GEOS-Chem grid configuration (i.e. 4° x 5°, 2° x 2.5°, and 0.5° x 0.666° nested grids, etc.). The lai*.global files are read from disk by routines readlai.F and rdlai.F, which save the LAI data into arrays named XLAI (in CMN_DEP_mod.F) and XYLAI (in CMN_VEL_mod.F).
The XLAI and XYLAI arrays store the leaf area index as a function of Olson land type (cf Olson 1992 land map). The XLAI array has dimensions (IIPAR,JJPAR,NTYPE), where IIPAR is the number of longitudes in the horizontal grid, JJPAR is the number of latitudes, and NTYPE is the number of individual Olson land types per each GEOS-Chem grid box. The XYLAI array contained the same data as in XYLAI, but for historical reasons was dimensioned with only one spatial dimension: (IIPAR*JJPAR,NTYPE).
Please note that there is a direct correspondence between the LAI data stored in the lai*.global files (and saved into XLAI and XYLAI arrays) and the Olson land map. Therefore, if you are using the Olson 1992 land map (native resolution 0.5° x 0.5°, default in GEOS-Chem), then you must also use LAI data at 0.5° x 0.5° native resolution. On the other hand, if you are using the Olson 2001 land map (native resolution 0.25° x 0.25°), then you must also use LAI data at 0.25° x 0.25° resolution.
GEOS-Chem users had the option to use either LAI data from the AVHRR satellite instrument or the MODIS satellite instrument. Individual sets of lai*.global files were prepared for both AVHRR LAI and MODIS LAI data. Users could select either AVHRR LAI or MODIS LAI with a switch in the input.geos file. Customary usage is to select the MODIS LAI data, for reasons explained HERE.
--Bob Y. 14:22, 12 April 2012 (EDT)
LAI used by MEGAN biogenic emissions
The MEGAN biogenic emissions module also relies on MODIS LAI data. Routine RDISOLAI (in lai_mod.F) reads MODIS LAI at 1° x 1° native resolution from files in binary punch format. The LAI data is regridded on-the-fly to GEOS-Chem resolution and stored in arrays named ISOLAI (daily LAI), PMISOLAI (LAI for previous month), MISOLAI (LAI for this month), and NMISOLAI (LAI for next month), all in lai_mod.F. The MEGAN module gets the MODIS LAI data directly from these arrays.
When modifications were made for the GEOS-5 0.5° x 0.666° nested grids, separate subroutines were added into lai_mod.f to read from LAI data sets that were specifically regridded to each 0.5° x 0.666° nested grid window (China/SE Asia, N. America, and Europe). This adds another layer of complexity on top of an somewhat already overburdened code.
--Bob Y. 14:22, 12 April 2012 (EDT)
LAI used by the mercury simulation
In the GEOS-Chem mercury simulation, the routine which computes Hg emission from soils (SOILEMIS, in module land_mercury_mod.F) uses the daily averaged LAI (stored in the ISOLAI, in lai_mod.F) in order to attenuate solar radiation. As mentioned above, this LAI data is first read in from disk at 1° x 1° resolution and then regridded to the current GEOS-Chem resolution.
--Bob Y. 14:22, 12 April 2012 (EDT)
Issues that needed to be resolved
These different sources of LAI data (i.e. one data set stored in the lai*.global files at GEOS-Chem resolution, one data stored in binary files at 1° x 1° native resolution, and the special data sets for the 0.5° x 0.666° nested grids) present an inconsistency that should be resolved as GEOS-Chem development continues to advance.
Furthermore, for the Grid-independent GEOS-Chem project we must migrate away from ASCII files (which prevent efficient interfacing with external GCMs). We also cannot assume any particular horizontal grid, since that is now to be specified by the external GCM. Therefore, we must instead read the LAI data from disk at the same native resolution, and from this, populate the arrays that are used by the dry deposition, soil NOx emissions, and MEGAN biogenic emissions modules.
Finally, to facilitate simulations at ultra-fine horizontal resolution, we will eventually adopt the Olson 2001 land map, which has a native resolution of 0.25° x 0.25°. We will also use an updated version of the MODIS LAI data at 0.25 x 0.25 resolution. We need a flexible mechanism that will allow us to switch between LAI data at different native horizontal resolutions.
--Bob Y. 14:22, 12 April 2012 (EDT)
Implementation of an updated LAI code
To resolve the issues discussed in the preceding section, the GEOS-Chem Support Team has created the following new GEOS-Chem modules:
- olson_landmap_mod.F90
- Used to read in the Olson land cover map directly from netCDF files at its native resolution (0.5° x 0.5° if using Olson 1992, 0.25° x 0.25° if using Olson 2001). This removes the need for obsolete ASCII files. Please see our Olson land map wiki page for a complete description.
- modis_lai_mod.F90
- This module replaces the existing GEOS-Chem module lai_mod.F. Using the Olson land map information supplied by olson_landmap_mod.F90, the routines in modis_lai_mod.F90 perform the following tasks:
- Read MODIS LAI data from netCDF files at its native resoluton (either 0.5° x 0.5° or 0.25° x 0.25°, depending on which Olson land map version is used)
- Regrid MODIS LAI data to the current GEOS-Chem resolution on-the-fly.
- Populate the XLAI array for backwards compatibility with the existing legacy Dry deposition and Soil NOx emissions modules.des.
- Populate LAI arrays that are needed for MEGAN biogenic emissions and the Mercury simulation.
- mapping_mod.F90
- Used to pass land type and regridding information from olson_landmap_mod.F90 to modis_lai_mod.F90.
For the MEGAN biogenic emissions and Mercury simulations, we have now replaced variables from lai_mod.F with the corresponding variables from modis_lai_mod.F90 as shown in this table:
| Variable | Type | Which stores this quantity | In this module | Is now replaced by | In this module |
|---|---|---|---|---|---|
| ISOLAI | array | Daily interpolated leaf area index [cm2 cm-2] | lai_mod.F | GC_LAI | modis_lai_mod.F90 |
| PMISOLAI | array | Previous month's leaf area index, as read from disk [cm2 cm-2] | lai_mod.F | GC_LAI_PM | modis_lai_mod.F90 |
| MISOLAI | array | Current month's leaf area index, as read from disk [cm2 cm-2] | lai_mod.F | GC_LAI_CM | modis_lai_mod.F90 |
| NMISOLAI | array | Next month's leaf area index, as read from disk [cm2 cm-2] | lai_mod.F | GC_LAI_NM | modis_lai_mod.F90 |
| DAYS_BTW_M | scalar | # of days between the start of 2 successive LAI months | lai_mod.F | DAYS_BTW_MON | modis_lai_mod.F90 |
We have also taken the opportunity to replace the obsolete XYLAI array with XLAI, which will allow us to retire the obsolete legacy code module CMN_VEL_mod.F. We will keep the XLAI array for compatibility with the dry deposition module for the foreseeable future (at least until the dry deposition module is modernized).
--Bob Y. 14:15, 12 April 2012 (EDT)
Validation
Using Olson 1992 land map
We have tested the implementation of the new LAI scheme with 1-month benchmarks. We used the Olson 1992 land map (which is the default in GEOS-Chem) and MODIS LAI data, both at 0.5° x 0.5° resolution. Here are our results:
We have confirmed that the updated LAI scheme generates identical XLAI arrays w/r/t the old scheme. This causes the Dry deposition and Soil NOx emissions codes to behave in precisely the same way as before.
Because we are now regridding from finer resolution than 1° x 1°, the LAI stored in arrays in modis_lai_mod.F90 (i.e. in arrays GC_LAI, GC_LAI_PM, GC_LAI_CM and GC_LAI_NM) differ slightly when compared to GEOS-Chem output using LAI that was regridded from 1° x 1°. In general, individual LAI values at a particular grid box will generally agree to the first 3 decimal places.
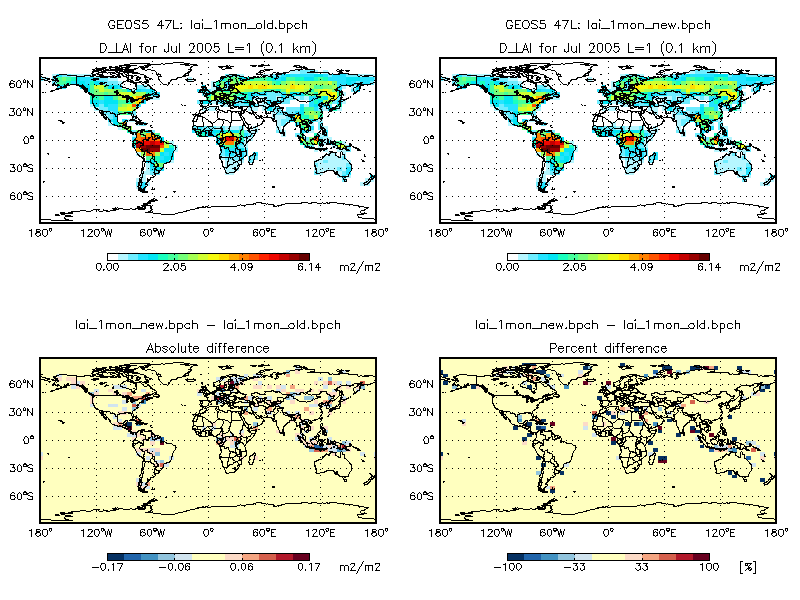
The plot below shows the monthly average of daily LAI (stored in the ISOLAI array of lai_mod.F) from v9-01-03i, to LAI computed by the updated scheme (stored in the GC_LAI array of modis_lai_mod.F90). As you can see, maximum difference for this month is less than 0.2 cm2 cm-2.
We have performed a 1-month benchmark (July 2005, using 4° x 5° GEOS-5 met data) using the new LAI scheme and have compared that to v9-01-03i. ISORROPIA was turned off in both simulations to negate the random numerical noise error. The following plots and tables show very minor differences in tracer concentrations and emissions. We attribute these differences to the fact that we are now regridding LAI from the native resolution of 0.5° x 0.5° instead of from 1° x 1°. This small numerical change causes further small differences, particularly in the MEGAN biogenic emissions module.
| Quantity being plotted | Plot | Remarks |
|---|---|---|
| Frequency distribution histogram | Click HERE to view | Most species have close to 100% of data points within the central bar. ISOP and its products have some spread, but that is likely due to small numerical differences. |
| Emissions table | Click HERE to view | NOTE: isoprene emissions |
| Emissions ratio for isoprene | Click HERE to view | For ISOP, the differences in ratio tend to occur where ISOP concentrations are already very small. |
| Absolute differences @ surface and 500hPa, all species (using fixed data ranges) | Click HERE to view | |
| Absolute differences @ surface and 500hPa, all species (using the dynamic range of the data) | Click HERE to view | Many species have abs diffs less than +/-0.05 ppbv. The largest differences are ISOP (+/-0.7 ppbC) and CO (+/-0.6 ppbv). These differences are likely well within the level of the uncertainties of the emissions data. |
| Zonal mean differences, all species (using fixed data ranges) | Click HERE to view | |
| Zonal mean differences, all species (using the dynamic range of the data) | Click HERE to view | All species show very small zonal mean differences, typically +/-0.2 ppbv or less. |
--Bob Y. 16:57, 12 April 2012 (EDT)
Using Olson 2001 land map
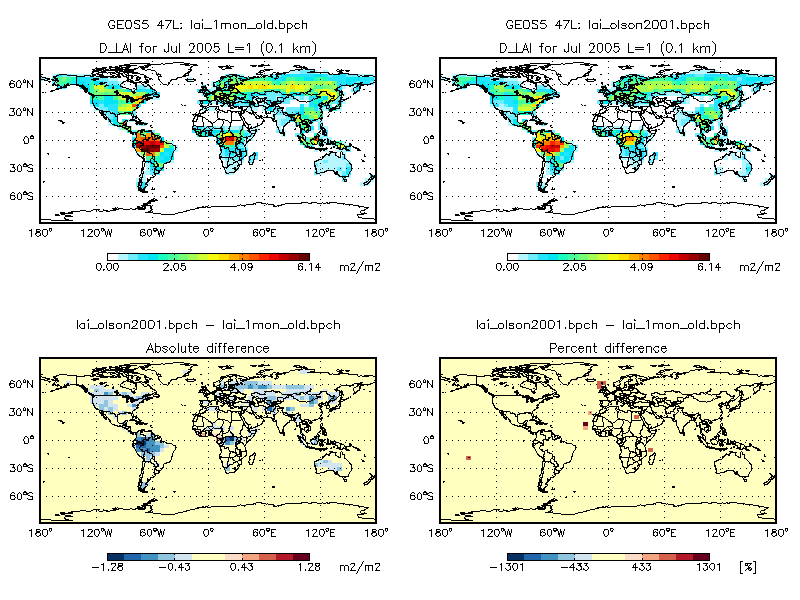
Switching from the Olson 1992 land map to the Olson 2001 land map seems to produce very different results in the GEOS-Chem full-chemistry simulation. The plot below shows the monthly average of daily-interpolated LAI values in a GEOS-Chem simulation with GEOS-5 met fields at 4° x 5° resolution. The left panel shows the monthly-mean LAI from v9-01-03i; the right panel is taken from a 1-month benchmark using the Olson 2001 land map and MODIS LAI (both at 0.25° x 0.25° native resolution). The absolute and percent differences are also plotted.
As you can see there is a marked discrepancy in the LAI values. The new LAI values over the Amazon lower by ~1 cm2 cm-2, and by approximately 0.5 cm2 cm-2 over much of North America.
It should be noted that the problem may not just be due to regridding from 0.25° x 0.25° to 4° x 5° resolution. The Olson 2001 land map uses a different land type numbering system than the Olson 1992 land map.
We have performed a 1-month benchmark (July 2005, using 4° x 5° GEOS-5 met data) using the updated LAI scheme with the Olson 2001 land map, and have compared that to v9-01-03i. ISORROPIA was turned off in both simulations to negate the random numerical noise error. The following plots and tables show very minor differences in tracer concentrations and emissions.
| Quantity being plotted | Plot | Remarks |
|---|---|---|
| Frequency distribution histogram | Click HERE to view | |
| Emissions table | Click HERE to view | NOTE: isoprene emissions |
| Emissions ratio for isoprene | Click HERE to view | |
| Absolute differences @ surface and 500hPa, all species (using fixed data ranges) | Click HERE to view | |
| Absolute differences @ surface and 500hPa, all species (using the dynamic range of the data) | Click HERE to view | |
| Zonal mean differences, all species (using fixed data ranges) | Click HERE to view | |
| Zonal mean differences, all species (using the dynamic range of the data) | Click HERE to view |