FAST-JX v7.0 photolysis mechanism: Difference between revisions
| Line 263: | Line 263: | ||
=== FAST-JX directory === | === FAST-JX directory === | ||
<span style="color:darkorange">'''''This update will be included in [[GEOS-Chem 12#12.4.0|GEOS-Chem 12.4.0]].'''''</span> | |||
In [[GEOS-Chem 12#12.4.0|GEOS-Chem 12.4.0]], the Photolysis Menu was introduced in <tt>input.geos</tt> for specifying the file path for the FAST-JX input files. This update was made to facilitate development of photolysis chemistry. | In [[GEOS-Chem 12#12.4.0|GEOS-Chem 12.4.0]], the Photolysis Menu was introduced in <tt>input.geos</tt> for specifying the file path for the FAST-JX input files. This update was made to facilitate development of photolysis chemistry. | ||
Revision as of 17:55, 30 April 2019
Sebastian Eastham (MIT) has introduced FAST-JX v7.0 into GEOS-Chem v10-01 concurrently with the UCX chemistry mechanism. FAST-JX v7.0 replaces the older FAST-J photolysis mechanism.
Overview
This update was validated in the 1-month benchmark simulation v10-01c and approved on 29 May 2014.
Sebastian Eastham incorporated Fast-JX v7.0a into the GEOS-Chem UCX mechanism. From Eastham et al. (2014):
- GEOS-Chem uses a customized version of the FAST-JX v6.2 photolysis mechanism (Wild et al., 2000), which efficiently estimates tropospheric photolysis. The customized version uses the wavelength bands from the older Fast-J tropospheric photolysis scheme and does not consider wavelengths shorter than 289 nm, assuming they are attenuated above the tropopause. However, these high-energy photons are responsible for the release of ozone-depleting agents in the stratosphere. The standard Fast-JX model (Prather, 2012) addresses this limitation by expanding the spectrum analyzed to 18 wavelength bins covering 177–850 nm, extending the upper altitude limit to approximately 60 km. We therefore incorporate Fast-JX v7.0a into GEOS-Chem UCX. Fast-JX includes cross-section data for many species relevant to the troposphere and stratosphere. However, accurately representing sulfur requires calculation of gaseous H2SO4 photolysis, a reaction which is not present in Fast-JX but which acts as a source of sulfur dioxide in the upper stratosphere. Based on a study by Mills (2005), the mean cross-section between 412.5 and 850 nm is estimated at 2.542 × 10−25 cm2. We also add photolysis of ClOO and ClNO2, given their importance in catalytic ozone destruction, using data from JPL 10-06 (Sander et al., 2011). Fast-JX v7.0a includes a correction to calculated acetone cross sections. Accordingly, where hydroxyacetone cross-sections were previously estimated based on one branch of the acetone decomposition, a distinct set of cross sections from JPL 10-06 are used.
- The base version of GEOS-Chem uses satellite observations of total ozone columns when determining ozone-related scattering and extinction. The UCX allows either this approach, as was used for the production of the results shown, or can employ calculated ozone mixing ratios instead, allowing photolysis rates to respond to changes in the stratospheric ozone layer.
Timeline
The following table displays a timeline of important milestones in FAST-JX v7.0 development:
| Version | Date | Features / Improvements |
|---|---|---|
| GEOS-Chem v10-01 | Jun 2015 |
|
--Bob Y. 13:42, 21 May 2014 (EDT)
Input files for FAST-JX v7.0
The following input files are required for the FAST-JX v7.0 photolysis mechanism:
| File | Introduced | Retired | Description |
|---|---|---|---|
| fastj.jv_atms.dat.nc | v9-01-03 | still used | Purpose:
Location:
NOTE: Where such data exists, GEOS-Chem will overwrite the reference O3 climatology as follows: |
| FJX_j2j.dat | v10-01c | still used | Purpose:
Location:
Replaces the following files:
|
| FJX_spec.dat | v10-01c | still used | Purpose
Location:
Replaces the following files:
|
| jv_spec.mie.dat | v10-01c | still used | Purpose:
Location:
Notes:
|
| brc.dat | v11-01 | still used | Purpose:
Location:
|
| dust.dat | v10-01 | still used | Purpose:
Location:
|
| org.dat | v10-01i | still used | Purpose:
Location:
|
| so4.dat | v10-01i | still used | Purpose:
Location:
|
| soot.dat | v10-01i | still used | Purpose:
Location:
|
| ssa.dat | v10-01i | still used | Purpose:
Location:
|
| ssc.dat | v10-01i | still used | Purpose:
Location:
|
| h2so4.dat | v10-01 public comment period | still used | Purpose:
Location:
|
| Files that have since been retired | |||
| jv_spec_aod.dat | v9-01-03 | v10-01i | Purpose:
|
--Melissa Sulprizio (talk) 12:35, 19 September 2018 (UTC)
--Bob Yantosca (talk) 20:15, 10 April 2019 (UTC)
FJX_j2j.dat
Seb Eastham wrote:
Each row in FJX_j2j.dat can be broken down as follows. Taking the ETP photolysis reaction as an example:
80 ETP PHOTON OH HO2 ALD2 0.500 /CH3OOH/
The elements mean the following in terms of how the code parses them:
- 80: This is the internal index used by Fast-JX. When setting up the reaction in globchem.eqn, this is the number you need when indexing PHOTOL.
- ETP: This is the GEOS-Chem species that will undergo photolysis.
- PHOTON: This just makes clear that the reaction is photolysis.
- 0.500: This is the quantum yield of the reaction. Specifically, it is a flat multiplier applied to the first-order rate returned by Fast-JX for this reaction specifically.
- CH3OOH: This is the cross-section (from FJX_spec.dat) which will be used to calculate the first-order reaction rate.
As you can see, the product list is not used – in theory you could put anything here! However, it is very helpful for other users if the product list is correctly specified, so that they can cross check between FJX_j2j.dat and globchem.eqn (where the product list must be correct). The actual calculation order would go as follows:
- Fast-JX calculates the cross section data for CH3OOH, then applies the local actinic flux in the grid box to derive a first-order reaction rate. Let’s call this rate "R".
- Every reaction listed in FJX_j2j.dat which uses the "CH3OOH" cross section in the final column would take the rate "R" as its rate of reaction, and then multiply it by the quantum yield. In the case of ETP, we end up with an overall photolysis rate of 0.500*R.
- The rate is stored in PHOTOL based on the first column of the FJX_j2j.dat entry, specifically PHOTOL(80) = 0.500*R
- When performing chemistry, KPP will retrieve the reaction rate from PHOTOL(80) (see reaction 544 in the standard globchem.eqn file). It ignores the "hv" reactant, yielding a final loss rate for ETP + hv of: d[ETP]/dt = -0.500 * R * [ETP].
The short version of all this is:
- The stoichiometric coefficients do not need to be included in FJX_j2j.dat, but it helps for clarity
- Only the globchem.eqn products are important in terms of the calculation, but again having the products right in FJX_j2j.dat does help in terms of clarity
--Melissa Sulprizio (talk) 14:51, 6 March 2017 (UTC)
IMPORTANT NOTE: In GEOS-Chem 12.3.2 and later versions, a duplicate entry for ClNO2 was removed from FJX_j2j.dat.
--Bob Yantosca (talk) 20:59, 10 April 2019 (UTC)
FAST-JX directory
This update will be included in GEOS-Chem 12.4.0.
In GEOS-Chem 12.4.0, the Photolysis Menu was introduced in input.geos for specifying the file path for the FAST-JX input files. This update was made to facilitate development of photolysis chemistry.
Tomas Sherwen wrote:
- Use the directory in input.geos' photolysis menu as the directory to look for FAST-JX data files. The default setting is to use the files in /ExtData/CHEM_INPUTS/FAST_JX/ for most recent version (e.g. subfolder v2018-09). However, a user can now set the FAST-JX directory to be a directory of their choice (e.g. a model's run directory).
--Melissa Sulprizio (talk) 17:54, 30 April 2019 (UTC)
VOC photolysis in FAST-JX v7.0
These updates were validated in the 1-month benchmark simulation v10-01c and approved on 29 May 2014.
In the table below, we summarize VOC photolysis in Fast-JX v7.0. We also invite you to view our Comparison of GEOS-Chem Photolysis Rates document prepared by Chris Chan Miller.
| Old reaction | New reaction | New rate | Note |
|---|---|---|---|
| CH2O = HO2 + HO2 + CO (channel a) | same | New cross sections leads to an increase by 10% to 20% | This increase is consistent with JPL 2010. |
| CH2O = H2 + CO (channel b) | same | increase by 6% to 15% | This increase is consistent with JPL 2010. |
| PAN = 0.6MCO3 + 0.6NO2 + 0.4MO2 + 0.4NO3 | PAN = 0.7MCO3 + 0.7NO2 + 0.3MO2 + 0.3NO3 | same | JPL2010 suggests two channels(only one channel in FastJX-v7.0), branching ratio follows JPL2010 |
| ALD2 = MO2 + HO2 + CO | ALD2 = 0.88MO2 + HO2 + 0.88CO + 0.12MCO3 | large discrepancy is found between obs and model (see Chris's slides), pressure dependence is probably needed. The cross section is now updated by Michael Prather. | |
| ALD2 = CH4 + CO | this channel is turned off | this channel is not included in FastJX-v7.0 | |
| RCHO = ETO2 + HO2 + CO | same | No pressure dependence is observed, according to JPL2010. | |
| MP = CH2O + HO2 + OH | same | change is small. | |
| GLYX = 0.5H2 + CO + 0.5CH2O + 0.5CO | GLYX = H2 + 2CO GLYX = CH2O + CO |
Pressure dependence is now included, rate is higher | now 3 channels, Stern-Volmer expression: Qtotal = 1/[6.80 + 251.8e-4 P(Torr)], Increases quantum yields at low P, but ONLY specified for 390-470 nm. Assume that 220K has P = 0.18 atm and higher q.(From FJX v7.0 notes) |
| GLYX = 2.0CO + 2.0HO2 | same | Pressure dependence is now included, rate is higher | |
| MACR = CO + HO2 + CH2O + MCO3 branching ratio = 0.5 |
same | rate is significantly reduced, as Qy is reduced from 0.008 to 0.003 | this channel is dominant,the third channel (C3H6 + CO), is ignored, to be consistent with Fast-JX v7.0 |
| MACR = MAO3 + HO2 branching ratio = 0.5 |
remove this channel | suggested by IUPAC, also consistent with Fast-JX v7.0 | |
| MVK = PRPE + CO branching ratio = 0.6 |
pressure dependence is now included | ||
| MVK = MCO3 + CH2O + CO + HO2 branching ratio = 0.2 |
pressure dependence is now included | ||
| MVK = MO2 + MAO3 branching ratio = 0.2 |
MVK = MO2 + RCO3 | this channel was removed in FJX v7.0, but it shouldn't according to IUPAC. MAO3 is changed to RCO3 for carbon balance. | |
| GLYC = CH2O + 2.0HO2 + CO | GLYC = 0.9CH2O + 1.73HO2 + 0.07OH + 1.0CO + 0.1MOH | Significant increase in X sections | merge from three channels GLYC =CH2O + 2.0HO2 + CO(QY = 0.83), GLYC =CH3OH + CO (QY=0.10), GLYC =OH + CH2O + HO2 + CO (QY =0.07) JPL 2010 |
| MEK = 0.85MCO3 + 0.85ETO2 + 0.15MO2 + 0.15RCO3 | same | two channels are merged into one reaction | |
| HAC = MCO3 + CH2O + HO2 | rate is lower | the old rate was using Acetone X sections.This species is not in FastJX v7.0. Seb added X sections based on JPL2010. Need to multiply 0.6 for quantum yield.The bins above 335nm must be zeroed out, otherwise J(HAC) would be too high. The major removal process is its reaction with OH, photolysis is of minor importance (see Orlando et al., 1999). |
--Jmao 15:54, 20 May 2014 (EDT)
Final recommendation for J(HAC) and J(PAN)
These updates were validated in the 1-month benchmark simulation v10-01d and approved on 03 Jun 2014.
Jingqiu Mao wrote:
- I have two more suggestions to the code and I think we then can finalize v10-01c. We can deal with unresolved J(VOC) later. Seb, please let me know if you think otherwise.
- For HAC, keep the QY as 0.6, but zero out the bins >335 nm.
- For PAN, change the reaction from
PAN = 0.6MCO3 + 0.6NO2 + 0.4MO2 + 0.4NO3
- to
PAN = 0.7MCO3 + 0.7NO2 + 0.3MO2 + 0.3NO3
Sebastian Eastham replied:
- These sound good to me, and I’m not aware of any other pressing issues regarding J-values.
Daniel Jacob replied:
- Thanks Jingqiu! If the 1-year benchmark run has already started just let it run - these changes will have very little effect except for HAC and we can just make a note of it. I'm glad that we resolved these J(VOC) issues thanks to Seb, Chris and Jingqiu. At this point we need to move on.
--Bob Y. 17:18, 30 May 2014 (EDT)
Overhead ozone columns for use with FAST-JX
Use online ozone in FAST-JX v7.0 instead of scaling ozone climatology to archived TO3 values
When using FAST-JX v7.0 with met data other than GEOS-5, we recommend that you select the following option in the CHEMISTRY MENU section of input.geos:
Online O3 for FAST-JX? : T
Selecting this option will cause FAST-JX v7.0 to copy the "online" O3 tracer concentration—contained in the State_Chm%TRACERS derived type object—directly into the FAST-JX module. O3 concentrations will be copied for all grid boxes starting at the surface and ending at the top of the chemistry grid, which is either the stratopause (for simulations using the UCX combined stratospheric-tropospheric chemistry mechanism) or the tropopause (for simulations not using UCX).
Using the online O3 option for FAST-JX v7.0 in conjunction with the UCX chemistry mechanism will allow photolysis rates to respond to the changes in the dynamically-evolving stratospheric ozone layer. This will result in a more accurate simulation.
Even if you are performing a tropospheric-only chemistry simulation—that is, not using the UCX mechanism—you should still use the online O3 option for FAST-JX v7.0. GEOS-Chem simulations using the FAST-JX online O3 option do not differ significantly from simulations where the internal FAST-JX ozone profiles are scaled to monthly-mean TO3 from either the TOMS/SBUV archive or the GMAO met fields. Sebastian Eastham writes:
[The online O3 option in FAST-JX v7.0] should only affect the impact of tropospheric ozone when comparing between [1-month benchmarks] v10-01b and v10-01c_trop, right? The stratospheric ozone estimate from the point of view of Fast-JX should be identical between v10-01b and v10-01c_trop regardless of the online ozone option. My personal thoughts are that leaving the option on should be fine as long as we trust the tropospheric ozone estimates, although it shouldn’t make much of a difference (and if it does that is probably something I should look into).
Of course, if you turn this option off (WHICH IS NOT RECOMMENDED):
Online O3 for FAST-JX? : F
then FAST-JX will scale its internal ozone profiles to TO3 data as described in this section of our FAST-J photolysis mechanism wiki page.
Use TOMS ozone for all years when running simulations with GEOS-5
NOTE: This update will be added to GEOS-Chem v11-01.
Jenny Fisher wrote:
I have been doing some test runs of the tropchem simulation using v11-01f (I know this isn’t recommended, but it was what I had easy access to and I figured I might as well trial it since the 1-year benchmarks have already been done). I was running 2010-2011 — which requires GEOS-5 — and noticed a weird behaviour in the log files: global mean OH jumps up by nearly 50% between December 2010 and Jan 2011 (see output from log files at the end of this email). I haven’t looked super carefully, but I have a hunch that this is because TOMS O3 availability ends after 2010, and starting in 2011 the code therefore uses the O3 column from the met files instead (and I see this switch in the log files). I know it makes sense to use the met field O3 in GEOS-FP, but has anyone looked carefully at the O3 columns in GEOS-5? Given GEOS-5 only goes through the end of 2012 (and GEOS-FP doesn’t start until partway through 2012), I think it would be safer to treat all GEOS-5 runs the same with respect to O3 columns. Even if this isn’t responsible for the OH jump, it would ensure continuity for people running multi-year GEOS-5 simulations, which is still pretty common, especially for interpreting existing observational records. This shouldn’t be hard to do — looks like the TOMS O3 data files have been updated through 2015: http://acdb-ext.gsfc.nasa.gov/Data_services/merged/
Bob Yantosca replied:
Jenny, you had asked about extending the TOMS data past 2010. I took a quick look at the data on the website http://acdb-ext.gsfc.nasa.gov/Data_services/merged/index.html, but I couldn’t find a lat-lon gridded product (like what we have used in the past). Maybe it’s there and I’m not looking in the right folder.
Jenny Fisher replied:
Hmmm, you’re right - I can’t find that version either… All I see are 5° zonal means. I guess that will have to wait for now.
--Bob Yantosca (talk) 21:32, 3 November 2016 (UTC)
Fix for TOMS to address strange cycle in OH output
This update was included in v11-01j and approved on 03 Dec 2016
To fix this issue, Barron Henderson has reprocessed the TOMS/SBUV data files that are stored in the ExtData/HEMCO/TOMS_SBUV/v2015-03 data directory folder. The GEOS-Chem Support Team has now stored these reprocessed files in a separate folder ExtData/HEMCO/TOMS_SBUV/v2016-11/.
The updated version of GeosCore/toms_mod.F mentioned below has been merged into v11-01j, and has been added to the GEOS-Chem repository.
The discussion regarding this issue follows below.
Ben Brown-Steiner wrote:
I've been running GEOS-Chem v10 "out of the box" (geos5_2x25_tropchem) and started to look at the daily output via the ND50 diagnostic (although this also happens with daily output to the main file). More that one person here at MIT has found the same behavior within their own simulations, so I'm pretty sure it's not just me.
Monthly averages look fine, and daily averages follow the long-term monthly behavior, but with a very weird cycle. At the 15th of each month, something just...stops. Certain species start to decay (e.g. OH) while others spike (e.g. O3).
I've attached a few files demonstrating the strange behavior.
First is global surface O3 (this is every day for a year, January - December, the plotted dates are wonky, please ignore).
Next is surface OH, with a similar strange behavior.
I've also plotted POX at the surface and in the upper atmosphere.
It seems that this happens most prominently over the oceans that over the land, and influences almost every tracer (NO, CO, PRPE, ISOP).
We don't know what's going on, but we suspect that it's not an emissions problem (as a similar pattern exists in the upper atmosphere). Perhaps something with radiation?
Do you know what's going on? Or point me to someone who does?
Barron Henderson responded:
Got it and I think there is an easy fix. (@Ben: see test below)
After reviewing HEMCO/Core/hco_calc_mod.F90, I agree that this is almost certainly a HEMCO/TOMS compatibility issue. HEMCO identifies missing values based on the HCO_MISSVAL (hco_calc_mod.F90:849-856). All masked values are returned as 0 (hco_calc_mod.F90:1534-1536). However, I think that the TOMS NetCDF files are not masked appropriately.
The TOMS netcdf files use -999 as the "missing_value" and "_FillValue" parameter, but... the actual missing values are set to -999.99. As a result, these values are not getting masked.
Since the -999.99 values are not masked, they are used in the aggregation to a new resolution. The result is a value slightly higher than -999 and therefore not caught in toms_mod (toms_mod.F:418-426)
The solution is quite simple. We just update the "missing_value" and "_FillValue" in the netcdf file. This will change the way the NetCDF library passes data to HEMCO.
Updating the properties can be done with ncatted from nco with the command below.
ncatted -a _FillValue,DTOMS2,o,f,-999.99 -a missing_value,DTOMS2,o,f,-999.99 -a _FillValue,DTOMS1,o,f,-999.99 -a missing_value,DTOMS1,o,f,-999.99 -a _FillValue,TOMS,o,f,-999.99 -a missing_value,TOMS,o,f,-999.99 /path/to/yourfiles/ExtData/HEMCO/TOMS_SBUV/v2015-03/TOMS_O3col_2004.geos.1x1.nc
I have tested that ncdump correctly identifies the values as masked after the update, but I have not seen if HEMCO does. @Ben - Can you try this simple fix? Just update the path and run the command above for 2003-2006 (if you don't have NCO, I can make the edits and post a file for you). Then, re-run 2004.
Jenny Fisher wrote:
Thanks for the solution!! Looking at this, I wonder if there is also going to be a problem associated with these lines:
IF ( TOMS(I,J) > -999e+0_f4 .AND. & DTOMS2(I,J) > -999e+0_f4 ) THEN
- I took a quick look back at the code, it looks as though by default we set the ozone column to 0 then overwrite if there is data. I assume having zero overhead ozone column could be a problem…
- Should we set to a more sensible default value?
:I had this issue with the Hg simulation and created the files with long-term mean and present day mean values. But they still have missing values at the poles in winter, so a default value might be a better option.
Ben Brown-Steiner wrote:
I've changed the "_FillValue" and "missing_value" for all the TOMS datasets, but am still getting the identical cycles.
- I copied over the TOMS_O3col* files and made the changes, and redirected the HEMCO config file to use the altered files. Should I have done something else?
Barron Henderson responded:
I am proposing a fix for the TOMS/HEMCO issue that tries to address the immediate issue and the unobserved problem. Karl Seltzer tested the fix. I also want to note that the same problem may be in other parts of the system as well.
The attached figures show J(O3) for the US (old_new_us.png, 20-60N and 110W-60W) and the world (old_new_global.png) as a function of time. Note the drastically improved pattern and higher average for the world. The higher average for the world is due to using a fill value for the unobserved poles.
This is a three step fix: (1) process TOMS files; (2) prepare HEMCO rc for new TOMS; (3) update toms module. The steps are described below and supplemented by the attachments. I welcome input on the proposed fix, particularly with respect to the "fill value" approach.
STEP 1 - Reprocess TOMS
- (processing: toms2edges.txt; result figure: TOMS_O3col_2004.a.geos.1x1.TOMS1.png)
- 1) Fixes missing flag.
- 2) Switches from monthly mean (center) and two delta's to two edges.
- 3) Fills data forward in time assuming missing data is best represented by the last value available.
- 4) Any cells with no preceding valid value (e.g., January) with the annual mean.
- 5) Filling missing latitude to address missing data. (See below)
- a) I am making a bad assumption as a quick fix/proof of concept. I am using the South pole values as a surrogate for missing north pole values -- matching season with a simple 6 month offset. If both the north and south are missing (e.g., 2003), fill the data poleward.
- b) This can be replaced with a better fill value later.
STEP 2 - Edit 1 line and add 2 to HEMCO rc
Edit the TOMS input line to add ".a" after the year
Was:
* TOMS_O3_COL $ROOT/TOMS_SBUV/v2015-03/TOMS_O3col_$YYYY.geos.1x1.nc TOMS 1971-2010/1-12/1/0 C xy dobsons * - 1 1
Would be
* TOMS_O3_COL $ROOT/TOMS_SBUV/v2016-11/TOMS_O3col_$YYYY.a.geos.1x1.nc TOMS 1971-2010/1-12/1/0 C xy dobsons * - 1 1
Additions:
* TOMS1_O3_COL - TOMS1 - - - dobsons/day * - 1 1
* TOMS2_O3_COL - TOMS2 - - - dobsons/day * - 1 1
STEP 3 - Replace toms_mod.F with the attached version. Recompile and run.
--Melissa Sulprizio (talk) 13:51, 21 July 2016 (UTC)
--Bob Yantosca (talk) 21:37, 3 November 2016 (UTC)
Cloud overlap options in FAST-JX v7.0
You may use the following cloud overlap options with the FAST-JX v7.0 photolysis mechanism:
Approximate random overlap assumption
The approximate random overlap option (which is the default setting) is:
Grid Box Optical Depth = In-Cloud Optical Depth * ( Cloud Fraction )^1.5
To select this option, make sure the following lines at the top of GeosCore/fast_jx_mod.F are uncommented:
! Approximate random overlap (balance between accuracy & speed) #define USE_APPROX_RANDOM_OVERLAP 1
As this is the default option, these lines should already be uncommented for you when you download the GEOS-Chem source code.
--Bob Y. 11:49, 20 May 2014 (EDT)
Linear cloud overlap assumption
The linear cloud overlap option is:
Grid Box Optical depth = In-cloud optical depth * Cloud fraction.
To select this option you must uncomment these lines at the top of GeosCore/fast_jx_mod.F:
!! Linear overlap !#define USE_LINEAR_OVERLAP 1
and then recompile GEOS-Chem.
--Bob Y. 11:49, 20 May 2014 (EDT)
Maximum random overlap assumption
At present, the maximum random overlap assumption has not been implemented into FAST-JX v7.0. Because this option is computationally intensive, it remains a research option rather than a standard supported feature.
--Bob Y. 11:49, 20 May 2014 (EDT)
Discussion
We invite you to read this discussion about cloud overlap options on our FAST-J photolysis mechanism wiki page.
--Bob Y. 16:13, 20 May 2014 (EDT)
Aerosol optical properties in FAST-JX v7.0
The aerosol optical properties have been updated from the older FAST-J photolysis mechanism. TEXT NEEDED
The ability to scale aerosol optical depth diagnostic output from 550 nm to other wavelengths (originally implemented for the older FAST-J mechanism in GEOS-Chem v8-03-01) is still compatible for FAST-JX v7.0. Please see this section on our FAST-J photolysis mechanism wiki page for more information.
--Bob Y. 11:12, 20 May 2014 (EDT)
Previous issues that have now been resolved
In this section we discuss issues that have been recently fixed in the implementation of FAST-JX v7.0:
Reactivation of bromine species photolysis for tropospheric simulation
This update was validated in the 1-month benchmark simulation v10-01c and approved on 29 May 2014.
Sebastian Eastham wrote:
- Bromine species photolysis should probably be reactivated in the tropospheric version – given that it was online in the pre-UCX version, we may as well keep it online. Doing so is just a question of removing the 'x' in the FJX_spec.dat file for the relevant species.
In FJX_spec.dat change the following lines from:
BrO x300 0.000E+00 0.000E+00 0.000E+00 0.000E+00 0.000E+00 0.000E+00 J10
0.000E+00 0.000E+00 0.000E+00 5.620E-19 1.202E-18 2.008E-18
3.239E-18 4.520E-18 5.064E-18 5.809E-18 7.350E-19 0.000E+00
BrNO3 x200 0.000E+00 0.000E+00 5.484E-19 7.245E-19 3.702E-18 3.475E-18 J10
3.182E-18 2.978E-18 5.304E-19 6.086E-19 4.489E-19 1.963E-19
1.584E-19 1.307E-19 1.110E-19 8.033E-20 3.377E-20 1.270E-21
BrNO3 x300 0.000E+00 0.000E+00 8.026E-19 1.071E-18 5.166E-18 4.190E-18 J10
3.467E-18 3.039E-18 5.567E-19 5.989E-19 4.528E-19 2.098E-19
1.705E-19 1.425E-19 1.207E-19 8.648E-20 3.716E-20 1.445E-21
HOBr x300 0.000E+00 0.000E+00 0.000E+00 0.000E+00 0.000E+00 0.000E+00 J10
0.000E+00 0.000E+00 1.324E-19 2.011E-19 2.202E-19 2.196E-19
1.726E-19 1.367E-19 1.157E-19 1.125E-19 6.197E-20 2.755E-21
to:
BrO 300 0.000E+00 0.000E+00 0.000E+00 0.000E+00 0.000E+00 0.000E+00 J10
0.000E+00 0.000E+00 0.000E+00 5.620E-19 1.202E-18 2.008E-18
3.239E-18 4.520E-18 5.064E-18 5.809E-18 7.350E-19 0.000E+00
BrNO3 200 0.000E+00 0.000E+00 5.484E-19 7.245E-19 3.702E-18 3.475E-18 J10
3.182E-18 2.978E-18 5.304E-19 6.086E-19 4.489E-19 1.963E-19
1.584E-19 1.307E-19 1.110E-19 8.033E-20 3.377E-20 1.270E-21
BrNO3 300 0.000E+00 0.000E+00 8.026E-19 1.071E-18 5.166E-18 4.190E-18 J10
3.467E-18 3.039E-18 5.567E-19 5.989E-19 4.528E-19 2.098E-19
1.705E-19 1.425E-19 1.207E-19 8.648E-20 3.716E-20 1.445E-21
HOBr 300 0.000E+00 0.000E+00 0.000E+00 0.000E+00 0.000E+00 0.000E+00 J10
0.000E+00 0.000E+00 1.324E-19 2.011E-19 2.202E-19 2.196E-19
1.726E-19 1.367E-19 1.157E-19 1.125E-19 6.197E-20 2.755E-21
--Melissa Sulprizio 13:33, 14 May 2014 (EDT)
Error in reducing wavelength bins for tropospheric simulation
This update was validated in the 1-month benchmark simulation v10-01c and approved on 29 May 2014.
Sebastian Eastham wrote:
- In fast_jx_mod, specifically RD_XXX, there is a transformation to reduce 18 cross sections to 12. Since bin 18 now corresponds to bin 12 and so on, the wavelengths are moved within the cross section array QQQ. However, the 12-bin capability is rarely used (if ever), so when Fast-JX was extended to allow cross sections with 1 or 3 sets of data, the 12 and 8 bin codes were not updated accordingly. This results in very large cross sections for acetone at long wavelengths, because the shorter wavelength data is being used instead.
- I've notified Michael Prather - he did not know about this bug and is putting together a fix ASAP. I've written my own fix in the meantime, which results in the acetone cross sections matching much more closely, at least between the two v10-01c versions.
--Melissa Sulprizio 10:39, 12 May 2014 (EDT)
Use proper org.dat file with updates for OC growth
This update was added to the official release of GEOS-Chem v10-01 (approved 15 Jun 2015).
After the GEOS-Chem v10-01i benchmarks were submitted, we learned that we were using a version of the FAST-JX input file org.dat that did not have Randall Martin's updates for better representation of OC growth with RH.
David Ridley wrote:
So it looks like I’d assumed you didn’t have the organics updates yet, so the org.dat I sent you doesn’t include Randall’s updates in there, sorry! I do have the multi-wavelength version that does include those. The change in the org optics seems to tie in with the reduction in the organic AOD we’re seeing, so that makes sense.
The BC optics are the same, so the differences seen there shouldn’t be coming from the optics. There may have been a change in the density because of a lot of BC tweaks around the same time. I’ll double check that.
We will make sure that the updated org.dat file ships with the publicly-released version of GEOS-Chem v10-01.
--Melissa Sulprizio 15:53, 11 May 2015 (EDT)
Unresolved issues
The following are active areas of research in GEOS-Chem, or ar issues that have not yet been resolved.
Duplicate entries for species ClNO2 in FJX_j2j.dat file
This issue is slated to be resolved in GEOS-Chem 12.3.2.
Bob Yantosca wrote:
I have found a mismatch in ClNO2 when comparing the noontime J-value diagnostics that are archived to bpch format vs. those archived to netCDF format. I believe that this issue is caused by the fact that ClNO2 is listed twice in the FAST-JX input file FJX_j2j.dat:
21 ClNO2 PHOTON Cl NO2 1.000 /ClNO2 /
. . .
101 ClNO2 PHOTON Cl NO2 1.000 /ClNO2 /
It seems like this might be a duplication, as both entries are mapped to the same FAST-JX species, and both entries have branching ratios of 1.0. (Usually if you list the same species more than once, they would have different branching ratios.)
The bpch diagnostic is hardwired to use the index, and not the name. But it is written only to take the first ClNO2 entry (#21) and to ignore the second entry (#101):
AD22(ILON,ILAT,L,47) = AD22(ILON,ILAT,L,47) + ! JClNO2
& ZPJ(L,21,ILON,ILAT)
But I think that the netCDF diagnostic seems to be picking up both ClNO2 entries #21 and #101.
Solution:
To fix this issue in GEOS-Chem 12.3.2 we have created a new folder in the GEOS-Chem shared data directories:
ExtData/CHEM_INPUTS/FAST_JX/v2019-04
which is a copy of the former FAST-JX input file folder (ExtData/CHEM_INPUTS/FAST_JX/v2018-09).
We edited the v2019-04/FJX_j2j.dat file so as to remove the duplicate entry (#101) for the ClNO2 species. As a result, the number of photolyzed GEOS-Chem species listed in v2019-04/FJX_j2j.dat was been reduced by 1, from 130 to 129. In order to be consistent with this update, in GEOS-Chem 12.3.2 and later versions, we have also set the parameter JVN_ = 129 in module Headers/CMN_FJX_MOD.F. The ND22 bpch diagnostic section in GeosCore/fast_jx_mod.F was also adjusted to account for the removal of the duplicate ClNO2 entry #101.
GEOS-Chem 12.3.2 and later versions now read the FAST-JX inputs from the v2019-04 folder as opposed to the former v2018-09 folder.
--Bob Yantosca (talk) 20:39, 10 April 2019 (UTC)
Species DHDC was not listed as photolyzing in the species database
This issue is slated to be resolved in GEOS-Chem 12.3.2.
Bob Yantosca wrote:
I've discovered an issue in photolysis for species DHDC (Dihydroxyperoxide dicarbonyl) in GEOS-Chem 12. There appears to be a typo in either the in either the FAST-JX input file FJX_j2j.dat, or in the species database. Long story short, in the FJX_j2j.dat file, we have an entry for species DHDC (Dihydroxyperoxide dicarbonyl) but not for DHDN:
109 DHDC PHOTON MGLY GLYX OH 1.000 /MeAcr /
But in the GEOS-Chem species database (defined in Headers/species_database_mod.F90), DHDN is listed as photolyzing, not DHDC:
CASE( 'DHDC' )
FullName = 'Dihydroxyperoxide dicarbonyl'
Formula = 'C5H8O6'
MW_g = 164.0_fp
Is_Gas = T
Is_Drydep = F
Is_Wetdep = F
CASE( 'DHDN' )
! DHDN uses the same DD_F0 and DD_Hstar_old values as ISOPN
! so that we can compute its drydep velocity explicitly.
FullName = 'C5 dihydroxydinitrate'
Formula = 'C5H10O8N2'
MW_g = 226.0_fp
Is_Gas = T
Is_Drydep = T
Is_Wetdep = T
Is_Photolysis = T
DD_F0 = 1.0_fp
DD_Hstar_Old = 2.00e+6_fp
Henry_K0 = 2.00e+6_f8
Henry_CR = 9200.0_f8
WD_RetFactor = 2.0e-2_fp
Kelvin Bates replied:
It looks from Chris Chan Miller's 2017 ACP paper like DHDC is definitely meant to photolyze; that was the source of some of the species he needed to match the isoprene products with observations. Those products are also the ones listed in the FJX_j2j.dat file, and the photolysis rate listed there (MeAcr) matches what I'd expect DHDC to have.
I can't find any literature on whether DHDN is supposed to photolyze, so I suspect it wasn't meant to. At least that means this should be an easy fix; we just need to add the Is_Photolysis = T line to the species database entry for DHDC and remove it from DHDN.
--Bob Yantosca (talk) 15:58, 10 April 2019 (UTC)
Carbonyl nitrate photolysis
Jingqiu Mao wrote:
- We know that carbonyl nitrate should be photolyzed much faster than the current rates in FAST-JX, according to this paper. But updates in this rate should also be combined with updates in the OH rate in order to better reproduce the experimental results from chamber studies. This seems more like a research question, so we decided to leave this to the SEAC4RS team.
--Bob Y. 15:40, 27 May 2014 (EDT)
Acetaldehyde photolysis
Jingqiu Mao wrote:
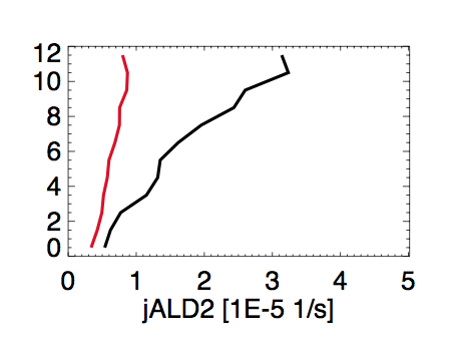
- We saw large discrepancies between observed(black) and modeled(red) J(ALD2), as shown in this plot by Chris Chan Miller:
- This discrepancy is very likely due to the lack of pressure dependence on the quantum yield. But Michael Prather didn’t include this pressure dependence in any of the FAST-JX versions. So this remains as a problem in all GEOS-Chem versions, including GEOS-Chem v10-01c.
Solution
These updates were validated with the 1-month benchmark simulation v10-01f and approved on Approved 13 Jan 2015.
The solution is to update the entries for acetaldehyde in the FAST-JX input file FJX_spec_dat, as follows:
Jingqiu Mao wrote:
- Michael Prather just provided a new set of cross section with pressure dependence for acetaldehyde:
ActAldp177 0.000E+00 0.000E+00 1.989E-23 0.000E+00 3.699E-22 4.938E-22 CH3CO IUPAC 2014
4.737E-22 4.659E-22 2.450E-20 3.409E-20 3.820E-20 3.732E-20 sheet P2 298K
2.707E-20 1.579E-20 6.566E-21 3.883E-22 5.683E-26 0.000E+00 q2=0.88 (CH3+HCO)
ActAldp566 0.000E+00 0.000E+00 1.903E-23 0.000E+00 3.539E-22 4.725E-22 q3=0.12 (H+CH3CO)
4.533E-22 4.458E-22 2.270E-20 2.985E-20 3.199E-20 2.987E-20 q1=0.00 (CH4+CO)
1.923E-20 9.497E-21 3.450E-21 1.914E-22 3.762E-26 0.000E+00 q's based on 1 bar
ActAldp999 0.000E+00 0.000E+00 1.822E-23 0.000E+00 3.389E-22 4.525E-22 wave > 300 nm
4.340E-22 4.269E-22 2.112E-20 2.647E-20 2.740E-20 2.479E-20
1.485E-20 6.739E-21 2.319E-21 1.258E-22 2.790E-26 0.000E+00
- We should use this instead. The reaction is also updated from
ALD2 = MO2 + HO2 + CO
- to
ALD2 = 0.88MO2 + HO2 + 0.88CO + 0.12MCO3
--Bob Y. 15:19, 6 June 2014 (EDT)
Validation
Eloise Marais wrote:
- I have implemented Michael Prather's pressure-dependent cross-sections for acetaldehyde (ALD2) in GEOS-Chem. The photolysis of ALD2 to form CH4 + CO is turned off. The product yields of the other ALD2 photolysis channel are also updated (see above). Pressure-dependent ALD2 photolysis leads to a decrease in J(ALD2) at the surface and an increase at 500 hPa. The effect on PAN is small (1-5 pptv increase at the surface and <2 pptv at 500 hPa in July 2005).
EP photolysis for dicarbonyls simulation
The SMVGEAR solver will be removed from GEOS-Chem v11-01, when the FlexChem solver package is implemented. At that time, routine calcrate.F will be removed from GEOS_Chem.
The code for EP photolysis found in calcrate.F needs to be updated for compatibility with FAST-JX v7.0. The EP photolysis code was left unchanged for now (as of GEOS-Chem v10-01c), but it is now executed only when LDICARB is true. This issue affects the dicarbonyls simulation.
!==============================================================
! HARDWIRE the effect of branching ratio of HOC2H4O in EP photolysis
! HOC2H4O ------> HO2 + 2CH2O : marked as I in P column of
! 'globchem.dat'
! HOC2H4O --O2--> HO2 + GLYC : marked as J in P column of
! 'globchem.dat'
!
! Add NCS index to NKHOROI and HKHOROJ for SMVGEARII (tmf, 12/16/06)
!==============================================================
! Not yet modified this for compatibility with Fast-JX v7.0.
! (SDE 04/01/13)
! Now only do the following if using the dicarbonyls mechanism
! (sde, mps, 5/28/14)
IF ( LDICARB ) THEN
IF ( NKHOROI(NCS) > 0 ) THEN
! Put J(EP) in correct spot for SMVGEAR II
PHOTVAL = NKHOROI(NCS) - NRATES(NCS)
NKN = NKNPHOTRT(PHOTVAL,NCS)
DO KLOOP=1,KTLOOP
RRATE(KLOOP,NKN)=RRATE(KLOOP,NKN) *
+ ( 1.D0-FYHORO(DENAIR(KLOOP), T3K(KLOOP)) )
ENDDO
ENDIF
IF ( NKHOROJ(NCS) > 0 ) THEN
! Put J(EP) in correct spot for SMVGEAR II
PHOTVAL = NKHOROJ(NCS) - NRATES(NCS)
NKN = NKNPHOTRT(PHOTVAL,NCS)
DO KLOOP=1,KTLOOP
RRATE(KLOOP,NKN)=RRATE(KLOOP,NKN) *
+ FYHORO(DENAIR(KLOOP), T3K(KLOOP))
ENDDO
ENDIF
ENDIF
--Melissa Sulprizio 13:35, 28 May 2014 (EDT)
References
- Blitz, M. A., D. E. Heard, M. J. Pilling, S. R. Arnold, M. P. Chipperfield, Pressure and temperature-dependent quantum yields for the photodissociation of acetone between 279 and 327.5 nm, Geophys. Res. Lett., 31, 9, L09104, 2004.
- Eastham, S. D., D. K. Weisenstein, S. R. H. Barrett, Development and evaluation of the unified tropospheric–stratospheric chemistry extension (UCX) for the global chemistry-transport model GEOS-Chem, Atmos. Environ, 89, 52-63, doi:10.1016/j.atmosenv.2014.02.001, 2014.
- Feng, Y., et al., Effects of cloud overlap in photochemical models, J. Geophys. Res., 109, D04310, doi:10.1029/2003JD004040, 2004.
- Liang, X.-Z., and W.-C. Wang, Cloud overlap effects on general circulation model climate simulations, J. Geophys. Res., 102 (D10), 11,039–11,047, 1997.
- Liu, H., et al., Radiative effect of clouds on tropospheric chemistry in a global three-dimensional chemical transport model, J. Geophys. Res., 111, D20303, doi:10.1029/2005JD006403, 2006.
- Magneron, I., A. Mellouki, G. Le Bras, G. K. Moortgat, A. Horowitz, and K. Wirtz , Photolysis and OH-Initiated Oxidation of Glycolaldehyde under Atmospheric Conditions, The Journal of Physical Chemistry A, 109(20), 4552-4561, doi:10.1021/jp044346y, 2005.
- Müller, J.-F., Peeters, J., and Stavrakou, T., Fast photolysis of carbonyl nitrates from isoprene, Atmos. Chem. Phys., 14, 2497-2508, doi:10.5194/acp-14-2497-2014, 2014.
- Orlando, J. J., G. S. Tyndall, J.-M. Fracheboud, E. G. Estupiñan, S. Haberkorn, and A. Zimmer, The rate and mechanism of the gas-phase oxidation of hydroxyacetone, Atmos. Environ., 33(10), 1621-1629, doi:10.1016/S1352-2310(98)00386-0,1999.
- Tie, X., et al., Effect of clouds on photolysis and oxidants in the troposphere, J. Geophys. Res., 108(D20), 4642, doi:10.1029/2003JD003659, 2003.
- Stubenrauch, C.J., et al., Implementation of subgrid cloud vertical structure inside a GCM and its effect on the radiation budget, J. Clim., 10, 273-287, 1997.
- Wild, O., X. Zhu, and M. J. Prather, Fast-J: Accurate simulation of in- and below-cloud photolysis in tropospheric chemical models, J. Atmos. Chem., 37, 245–282, 2000.
--Bob Y. 15:01, 27 May 2014 (EDT)