Profiling GEOS-Chem: Difference between revisions
| Line 130: | Line 130: | ||
=== Using PLOT_GPROF to create plots from the profiling data === | === Using PLOT_GPROF to create plots from the profiling data === | ||
You can use [http://acmg.seas.harvard.edu/gamap/ GAMAP] routine <tt>PLOT_GPROF</tt> to create a bar chart of the profiling results obtained from the <tt>gprof</tt> profiler. (NOTE: You will need to use [http://acmg.seas.harvard.edu/gamap/doc/index.html GAMAP v2-19 (release date: 15 Dec 2016)] or higher. | You can use [http://acmg.seas.harvard.edu/gamap/ GAMAP] routine <tt>PLOT_GPROF</tt> to create a bar chart of the profiling results obtained from the <tt>gprof</tt> profiler. (NOTE: You will need to use [http://acmg.seas.harvard.edu/gamap/doc/index.html GAMAP v2-19 (release date: 15 Dec 2016)] or higher.) <tt>PLOT_GPROF</tt> takes the following inputs: | ||
; CALLING SEQUENCE: | |||
; PLOT_GPROF, FILENAME [,keywords] | |||
; | |||
; INPUTS | |||
; FILENAME -> Text file containing profiling output from gprof. | |||
; | |||
; KEYWORD PARAMETERS: | |||
; N_DISPLAY -> The number of routines to display, from slowest | |||
; to fastest. The default is 30. | |||
; | |||
; BARCOLOR -> The color table value that will be used to plot | |||
; the bars. Default is !MYCT.RED. | |||
; | |||
; /VERBOSE -> If set, will print a listing of the N_DISPLAY | |||
; slowest routines. The routine name, execution time, | |||
; and percentage of total run time will be printed. | |||
To make a plot from the [[#Creating a table of time spent in each GEOS-Chem routine|the profiling output we generated in the previous section]], type at the IDL prompt: | |||
IDL> plot_gprof, 'GEOS-Chem_Profile_Results.txt' | |||
This will generate a plot that looks like this: | |||
[[Image:plot_gprof_output.png]] | [[Image:plot_gprof_output.png]] | ||
The length of the bar represents the time spent in each routine. The numbers printed to the right of each bar represent the percentage of the total execution time spent in that routine. | |||
If you would also like to print the profiling information, use the <tt>/VERBOSE</tt> keyword. Typing: | |||
IDL> plot_gprof, 'GEOS-Chem_Profile_Results.txt', /Verbose | |||
will create the plot above but also will print to the screen the time spent in each routine in seconds, plus the percentage of the total run time spent in each routine. | |||
The 30 slowest routines in profile: Profile_Gfortran_HJ.txt | |||
1 LMTPPM 720.110 s 15.860 % | |||
2 KPPDECOMP 679.390 s 14.960 % | |||
3 KPPSOLVE 451.530 s 9.940 % | |||
4 DO_REGRID_A2A 340.670 s 7.500 % | |||
5 FUN 264.420 s 5.820 % | |||
6 EMISSCO2 188.590 s 4.150 % | |||
7 RD_MIE 185.620 s 4.090 % | |||
8 JAC_SP 154.700 s 3.410 % | |||
9 EMISSIONS_INIT 148.260 s 3.270 % | |||
10 ROSENBROCK 146.630 s 3.230 % | |||
11 DO_CONVECTION 140.720 s 3.100 % | |||
12 VDINTI 133.510 s 2.940 % | |||
13 DO_RPMARES 126.700 s 2.790 % | |||
14 PHOTO_JX 112.290 s 2.470 % | |||
15 WAXPY 104.820 s 2.310 % | |||
16 SET_INITIAL_MIXRATIOS 99.390 s 2.190 % | |||
17 GEN_ID 44.030 s 0.970 % | |||
18 CALCACT4_ 43.530 s 0.960 % | |||
19 SET_HET 41.780 s 0.920 % | |||
20 GETIDX 37.350 s 0.820 % | |||
21 FINDFREELUN 25.320 s 0.560 % | |||
22 UNZIP_A6_FIELDS 23.130 s 0.510 % | |||
23 VDIFFDR 22.990 s 0.510 % | |||
24 GET_OH 22.290 s 0.490 % | |||
25 DO_DIAG_OH 17.220 s 0.380 % | |||
26 COMPUTE_F 16.820 s 0.370 % | |||
27 HCO_CALCEMIS 16.480 s 0.360 % | |||
28 SET_MINIT 16.460 s 0.360 % | |||
29 DIAG1_ 15.050 s 0.330 % | |||
30 GCJPLPR 14.080 s 0.310 % | |||
By default, <tt>PLOT_GPROF</tt> will show the 30 slowest routines. You can change the number of routines displayed by specifying a value with the <tt>N_DISPLAY</tt> keyword, e.g. | |||
IDL> plot_gprof, 'GEOS-Chem_Profile_Results.txt', N_DISPLAY=40 | |||
--[[User:Bmy|Bob Yantosca]] ([[User talk:Bmy|talk]]) 20:50, 15 December 2016 (UTC) | |||
=== For more information === | === For more information === | ||
Revision as of 20:50, 15 December 2016
On this page, we provide information on how you can use software profiling tools with GEOS-Chem.
Overview
Profiling is a debugging technique that allows you to examine how much time is spent in a given GEOS-Chem routine. This is helpful in identifying potential bottlenecks caused by sub-optimally written code. GEOS-Chem v11-01 and higher versions contains support for two software profiling packages:
The following sections describe how you can use GEOS-Chem with these utilities.
--Bob Yantosca (talk) 18:25, 14 December 2016 (UTC)
Profiling GEOS-Chem with gprof
The GNU profiler (gprof) is a free, open source software package that you can use to determine which GEOS-Chem routines take the most time to execute. If your computer system has the GNU Compiler Collection (GCC)] already installed (as do most modern Unix/Linux clusters), then gprof should also be available for you to use. GEOS-Chem v11-01 and higher versions contain support for gprof.
Compiling and running GEOS-Chem with gprof
To profile GEOS-Chem, you must first compile with the GPROF=y Makefile option, e.g.:
make -j4 GPROF=y ...etc. other makefile options ...
This will add the following compiler switches to the compilation sequence:
| Compiler | Switch to invoke gprof |
|---|---|
| Intel Fortran compiler (ifort) | -p |
| GNU Fortran compiler (gfortran) | -pg |
| PGI Fortran compiler (pgfortran) | -pg |
Once you have compiled GEOS-Chem with GPROF=y, you can run GEOS-Chem as you normally would. GEOS-Chem will create a binary file called gmon.out containing the profiling information. To generate readable output from gmon.out, you need to call the gprof program as described in the next section.
--Bob Yantosca (talk) 18:09, 14 December 2016 (UTC)
Generating profiling output
The general syntax for calling gprof is:
gprof [optional commands] [name of GEOS-Chem executable file] gmon.out > [name of text file to contain profiling results]
Please see this list of optional commands for gprof from the web-based documentation.
--Bob Yantosca (talk) 18:09, 14 December 2016 (UTC)
Creating a table of time spent in each GEOS-Chem routine
To create a simple text table of time spent in each GEOS-Chem subroutine (also called a flat profile), you can call gprof as follows:
gprof geos.mp gmon.out > GEOS-Chem_Profile_Results.txt
where geos.mp is the GEOS-Chem executable file. (It may have a different name than geos.mp depending on how, and in which run directory, you compiled GEOS-Chem.) You can also refine the output by specifying optional commands to gprof.
The GEOS-Chem_Profile_Results.txt file will contain output similar to this:
Flat profile:
Each sample counts as 0.01 seconds.
% cumulative self self total
time seconds seconds calls Ks/call Ks/call name
16.50 801.03 801.03 11317370 0.00 0.00 gckpp_linearalgebra_mp_kppdecomp_
9.63 1268.29 467.26 43751785 0.00 0.00 gckpp_linearalgebra_mp_kppsolve_
6.29 1573.53 305.24 26835672 0.00 0.00 gckpp_function_mp_fun_
4.09 1772.06 198.53 17402 0.00 0.00 tpcore_fvdas_mod_mp_fzppm_
4.04 1967.96 195.90 6834149 0.00 0.00 gckpp_jacobian_mp_jac_sp_
2.86 2106.99 139.03 72 0.00 0.04 flexchem_mod_mp_do_flexchem_
2.23 2215.23 108.25 182368327 0.00 0.00 gckpp_linearalgebra_mp_waxpy_
2.20 2322.10 106.87 45915 0.00 0.00 convection_mod_mp_do_merra_convection_
2.05 2421.82 99.72 648 0.00 0.00 dao_mod_mp_airqnt_
2.05 2521.43 99.61 3051 0.00 0.00 fast_jx_mod_mp_blkslv_
2.05 2620.81 99.38 3487 0.00 0.00 fast_jx_mod_mp_opmie_
1.80 2708.34 87.53 144 0.00 0.00 convection_mod_mp_do_convection_
1.78 2794.64 86.30 6533 0.00 0.00 vdiff_mod_mp_vdiff_
... etc ...
The information is arranged into several columns:
| Column | Description |
|---|---|
| % time | This is the percentage of the total execution time your program spent in this function. These should all add up to 100%. |
| cumulative seconds | This is the cumulative total number of seconds the computer spent executing this functions, plus the time spent in all the functions above this one in this table. |
| self seconds | This is the number of seconds accounted for by this function alone. The flat profile listing is sorted first by this number. |
| calls | This is the total number of times the function was called. If the function was never called, or the number of times it was called cannot be determined (probably because the function was not compiled with profiling enabled), the calls field is blank. |
| self ms/call | This represents the average number of milliseconds spent in this function per call, if this function is profiled. Otherwise, this field is blank for this function. |
| total ms/call | This represents the average number of milliseconds spent in this function and its descendants per call, if this function is profiled. Otherwise, this field is blank for this function. This is the only field in the flat profile that uses call graph analysis. |
| name | This is the name of the function. The flat profile is sorted by this field alphabetically after the self seconds and calls fields are sorted. |
Looking down the self seconds column will show you the amount of time spent in each routine. From the example above, we see that the first 3 routines (which are part of the KPP chemical solver) account for approximately one-third of the total run time.
--Bob Yantosca (talk) 18:19, 14 December 2016 (UTC)
Using PLOT_GPROF to create plots from the profiling data
You can use GAMAP routine PLOT_GPROF to create a bar chart of the profiling results obtained from the gprof profiler. (NOTE: You will need to use GAMAP v2-19 (release date: 15 Dec 2016) or higher.) PLOT_GPROF takes the following inputs:
; CALLING SEQUENCE: ; PLOT_GPROF, FILENAME [,keywords] ; ; INPUTS ; FILENAME -> Text file containing profiling output from gprof. ; ; KEYWORD PARAMETERS: ; N_DISPLAY -> The number of routines to display, from slowest ; to fastest. The default is 30. ; ; BARCOLOR -> The color table value that will be used to plot ; the bars. Default is !MYCT.RED. ; ; /VERBOSE -> If set, will print a listing of the N_DISPLAY ; slowest routines. The routine name, execution time, ; and percentage of total run time will be printed.
To make a plot from the the profiling output we generated in the previous section, type at the IDL prompt:
IDL> plot_gprof, 'GEOS-Chem_Profile_Results.txt'
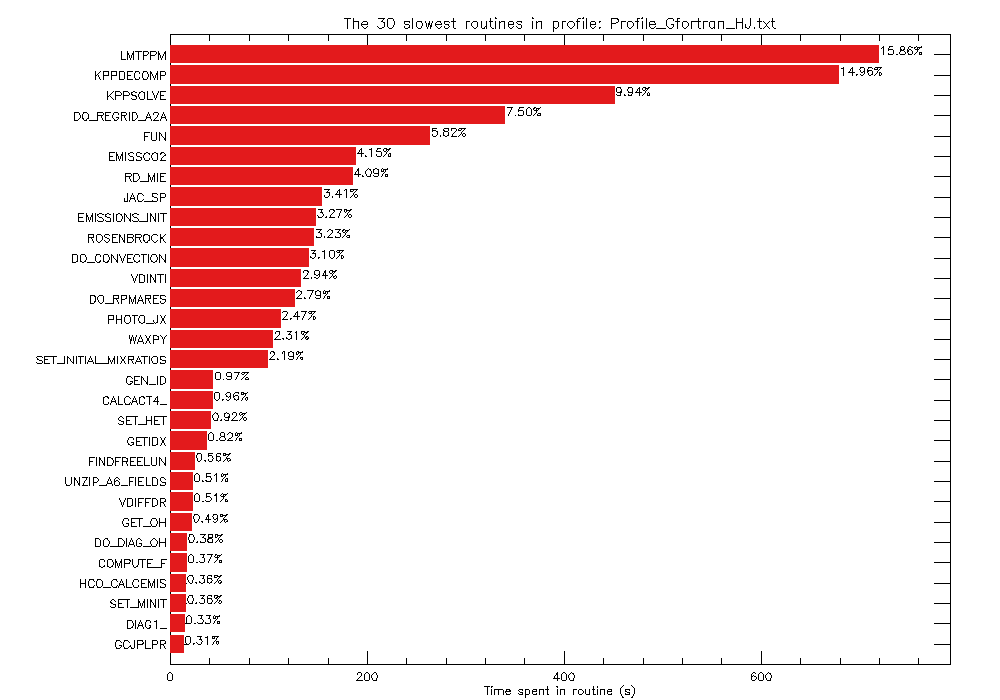
This will generate a plot that looks like this:
The length of the bar represents the time spent in each routine. The numbers printed to the right of each bar represent the percentage of the total execution time spent in that routine.
If you would also like to print the profiling information, use the /VERBOSE keyword. Typing:
IDL> plot_gprof, 'GEOS-Chem_Profile_Results.txt', /Verbose
will create the plot above but also will print to the screen the time spent in each routine in seconds, plus the percentage of the total run time spent in each routine.
The 30 slowest routines in profile: Profile_Gfortran_HJ.txt 1 LMTPPM 720.110 s 15.860 % 2 KPPDECOMP 679.390 s 14.960 % 3 KPPSOLVE 451.530 s 9.940 % 4 DO_REGRID_A2A 340.670 s 7.500 % 5 FUN 264.420 s 5.820 % 6 EMISSCO2 188.590 s 4.150 % 7 RD_MIE 185.620 s 4.090 % 8 JAC_SP 154.700 s 3.410 % 9 EMISSIONS_INIT 148.260 s 3.270 % 10 ROSENBROCK 146.630 s 3.230 % 11 DO_CONVECTION 140.720 s 3.100 % 12 VDINTI 133.510 s 2.940 % 13 DO_RPMARES 126.700 s 2.790 % 14 PHOTO_JX 112.290 s 2.470 % 15 WAXPY 104.820 s 2.310 % 16 SET_INITIAL_MIXRATIOS 99.390 s 2.190 % 17 GEN_ID 44.030 s 0.970 % 18 CALCACT4_ 43.530 s 0.960 % 19 SET_HET 41.780 s 0.920 % 20 GETIDX 37.350 s 0.820 % 21 FINDFREELUN 25.320 s 0.560 % 22 UNZIP_A6_FIELDS 23.130 s 0.510 % 23 VDIFFDR 22.990 s 0.510 % 24 GET_OH 22.290 s 0.490 % 25 DO_DIAG_OH 17.220 s 0.380 % 26 COMPUTE_F 16.820 s 0.370 % 27 HCO_CALCEMIS 16.480 s 0.360 % 28 SET_MINIT 16.460 s 0.360 % 29 DIAG1_ 15.050 s 0.330 % 30 GCJPLPR 14.080 s 0.310 %
By default, PLOT_GPROF will show the 30 slowest routines. You can change the number of routines displayed by specifying a value with the N_DISPLAY keyword, e.g.
IDL> plot_gprof, 'GEOS-Chem_Profile_Results.txt', N_DISPLAY=40
--Bob Yantosca (talk) 20:50, 15 December 2016 (UTC)
For more information
We invite you to read the gprof Manual to learn about other types of profiling output that you can generate with gprof.
--Bob Yantosca (talk) 18:30, 14 December 2016 (UTC)
Profiling GEOS-Chem with TAU
Information to be added