Issues now resolved in GEOS-Chem 12: Difference between revisions
| Line 89: | Line 89: | ||
=== Corrections for CFCs in the chemistry mechanism === | === Corrections for CFCs in the chemistry mechanism === | ||
<span style="color:green">'''''This update | <span style="color:green">'''''This update was included in [[GEOS-Chem_12#12.5.0|GEOS-Chem 12.5.0]], which was released on 09 Sep 2019.'''''</span> | ||
'''''Sebastian Eastham wrote:''''' | '''''Sebastian Eastham wrote:''''' | ||
Revision as of 16:45, 15 October 2019
The following bugs and/or technical issues have been resolved in GEOS-Chem 12.0.0 through GEOS-Chem 12.5.0. Issues resolved after GEOS-Chem 12.5.0 are listed as Closed on the following Github pages:
List of issues
Quick fix for aerosol pH calculation when dry
This update (Git ID: ce29c6c3) will be included in GEOS-Chem 12.6.0.
Xuan Wang wrote:
- My quick fix is very simple, which only changes one number in the source code:
- In GeosCore/ isorropiaII_mod.F, line 685, change
NITRTEMP = 1e-30_fp
State_Chm%pHSav(I,J,L) = -999e+0_fp
ELSE
- To
NITRTEMP = 1e-30_fp
State_Chm%pHSav(I,J,L) = 20e+0_fp
ELSE
Becky Alexander wrote:
- Note that [this fix] is not ideal. We have a better way of doing it which requires some sort of counting diagnostic. We have this working for .bpch only because in the version of the code that we are using (the one from Xuan's paper, v11-02d), not all of the netCDF diagnostics work. So we are unable to test this fix for the netCDF diagnostics.
--Melissa Sulprizio (talk) 17:13, 27 June 2019 (UTC)
GCHP silent bug when using gfortran compiler
This update was included in GEOS-Chem 12.5.0, which was released on 09 Sep 2019..
Compiling GCHP versions prior to 12.5.0 with GCC compiler gfortran results in silent bug where the Olson land mask is assigned all zeros. The issue has been isolated to the old version of MAPL that is replaced in GCHP 12.5.0, correcting the issue.
--Lizzie Lundgren (talk) 15:09, 26 April 2019 (UTC)
Enable accurate cloud pH diagnostic calculation
This update (Git ID: 1c15e54a) was included in GEOS-Chem 12.5.0, which was released on 09 Sep 2019.
Jonathan Moch implemented a new field State_Chm%Is_Cloud. To accurately calculate cloud pH in post-processing, you can divide Chem_pHCloud by Chem_isCloud. Both of these fields can be saved out from GEOS-Chem in HISTORY.rc via the State_Chm collection:
#==============================================================================
# %%%%% THE StateChm COLLECTION %%%%%
#
# Fields of the State_Chm object (see also Aerosols collection)
#
# Available for full-chemistry and aerosols-only simulations
#==============================================================================
StateChm.template: '%y4%m2%d2_%h2%n2z.nc4',
StateChm.format: 'CFIO',
StateChm.frequency: {FREQUENCY}
StateChm.duration: {DURATION}
StateChm.mode: 'time-averaged'
StateChm.fields: 'Chem_phSav ', 'GIGCchem',
'Chem_HplusSav ', 'GIGCchem',
'Chem_WaterSav ', 'GIGCchem',
'Chem_SulRatSav ', 'GIGCchem',
'Chem_NaRatSav ', 'GIGCchem',
'Chem_AcidPurSav ', 'GIGCchem',
'Chem_BiSulSav ', 'GIGCchem',
'Chem_pHCloud ', 'GIGCchem',
'Chem_isCloud ', 'GIGCchem',
'Chem_SSAlkAccum ', 'GIGCchem',
'Chem_SSAlkCoarse ', 'GIGCchem',
'Chem_HSO3AQ ', 'GIGCchem',
'Chem_SO3AQ ', 'GIGCchem',
'Chem_fupdateHOBr ', 'GIGCchem',
::
--Melissa Sulprizio (talk) 13:32, 27 June 2019 (UTC)
Fix typo in CLD_PARAMS routine in gckpp_HetRates.F90
This update (Git ID: 95d124e0) was included in GEOS-Chem 12.5.0, which was released on 09 Sep 2019.
Steve Steenrod wrote:
- I noticed line 5995 in KPP/Standard/gckpp_HetRates.F90 is:
IF (((QL.le.0.0e+0_fp).and.(QL.le.0.0e+0_fp)).or.(CLDF(I,J,L).le.0.0e+0_fp)) THEN
- An odd structure of "if ((A and A) or B)", so I was thinking one of the "QL"s should probably be "QI"
--Melissa Sulprizio (talk) 18:08, 18 June 2019 (UTC)
Corrections for CFCs in the chemistry mechanism
This update was included in GEOS-Chem 12.5.0, which was released on 09 Sep 2019.
Sebastian Eastham wrote:
- I’d like to recommend three changes to the standard chemistry mechanism for inclusion ASAP. These are basic science bug fixes, each of which was raised by Xuan Wang (cc’d). Each fix address a clear typo, where Cl is not being conserved in a stratospheric reaction. I have also sent them to the chemistry and stratospheric working groups. When I checked, each of these issues is still present in 12.4.0. The fixes:
- 1. Reaction of O1D with CFC12
O1D + CFC12 -> 0.14O + 0.14CFC12 + 0.86ClO + 1.72Cl
- should instead be
O1D + CFC12 -> 0.14O + 0.14CFC12 + 0.86ClO + 0.86Cl
- 2. Photolysis of HCFC142b and HCFC22
- These species only contain 1 Cl atom, but their photolysis in GC produces 2. The reactions should be changed from
HCFC142b + hv -> 2Cl HCFC22 + hv -> 2Cl
- to
HCFC142b + hv -> Cl HCFC22 + hv -> Cl
--Melissa Sulprizio (talk) 14:05, 17 June 2019 (UTC)
Retire obsolete emission inventories
This update was included in GEOS-Chem 12.5.0, which was released on 09 Sep 2019.
Eloise Marais wrote:
- My suggestion is to retire the following, as named in HEMCO_Config, so that users don’t mistakenly double-count and to make the process of selecting inventories easier for the user:
- BIOFUEL (outdated and all global inventories have this now)
- GEIA (all source types except natural NH3 emissions)
- BOND (same reason as for BIOFUEL)
- RETRO (would require an edit to HEMCO_Config so that if users select EDGAR for NOx, SOx, CO etc., CEDS VOC emissions are used)
- BRAVO (outdated and coarser resolution than the global inventories, so even though this is a regional inventory, it can’t be better now than EDGAR or CEDS)
- EMEP and EMEP_SHIP (same as for BRAVO. Mat Evans and I are working on processing the latest EMEP inventory to replace this one)
- TRASH (now included in CEDS, so redundant)
XIAO (this one I’m not 100% sure of. If this is still used by the community to address underestimates in other inventories, then ignore this suggestion).C2H6_2010 (same comment as for XIAO)
- [Trash burning is] not mentioned in the Crippa (2018) EDGAR v4.3.2 paper. Perhaps then if EDGAR is selected, it should include CEDS VOCs and Wiedinmyer (2014) TRASH for NOx, SOx, CO, BC and OC. We can be clear about this in the summary table on the wiki and the narrative description.
Melissa Sulprizio wrote:
- Here are the annual emissions for C2H6 and C3H8 using the different inventories:
C2H6 C2H8
Xiao 7.878592 11.88017 Tg
CEDS 6.538788 6.189306 Tg
Tzompa 7.044965 - Tg
Emily Fischer wrote:
- Given these maps and totals, I think we have to keep supporting Xiao and Tzompa. The CEDS totals are low, and it looks like the distribution is likely out of date. CEDS is going to make the model under-predict C2H6 somewhat, and C3H8 substantially compared to observations.
--Melissa Sulprizio (talk) 12:52, 14 June 2019 (UTC)
Bug fix for C3H8 anthropogenic emissions
This update was included in GEOS-Chem 12.5.0, which was released on 09 Sep 2019.
With the implementation of C2H6 emissions from Tzompa-Sosa et al. (2017) in GEOS-Chem v11-02f, C2H6 and C3H8 emissions from Xiao et al. (2008) were turned off. As a result, CEDS C3H8 emissions were used.
--> XIAO : false --> C2H6_2010 : true ... #============================================================================== # --- Xiao et al., JGR, 2008 --- # # NOTES: # - Xiao et al C2H6 and C3H8 are used in place of RETRO # See: http://wiki.geos-chem.org/Implementation_of_RETRO_Anthropogenic_Emissions#RETRO_anthropogenic_ethane_emissions_are_too_low # - Priority is set to 5 to overwrite RETRO, but not newer regional inventories (NEI2011, MIX) #============================================================================== (((XIAO 0 XIAO_C2H6 $ROOT/XIAO/v2014-09/C3H8_C2H6_ngas.geos.1x1.nc C2H6 1985/1/1/0 C xy kgC/m2/s C2H6 26 1 5 0 XIAO_C3H8 $ROOT/XIAO/v2014-09/C3H8_C2H6_ngas.geos.1x1.nc C3H8 1985/1/1/0 C xy kgC/m2/s C3H8 6/7/26/22 1 5 (((BIOFUEL 0 BIOFUEL_C2H6 $ROOT/BIOFUEL/v2014-07/biofuel.geos.4x5.nc BIOFUEL_C2H6 1985/1/1/0 C xy kgC/m2/s C2H6 - 2 5 0 BIOFUEL_C3H8 $ROOT/BIOFUEL/v2014-07/biofuel.geos.4x5.nc BIOFUEL_C3H8 1985/1/1/0 C xy kgC/m2/s C3H8 - 2 5 )))BIOFUEL )))XIAO #============================================================================== # --- 2010 Global fossil fuel and biofuel emissions of C2H6 (Tzompa-Sosa et al. (2017) --- #============================================================================== (((C2H6_2010 0 C2H6_2010_oilgas $ROOT/C2H6_2010/v2017-05/C2H6_global_anth_biof.2010$MM.nc ANTHR_C2H6 2010/1-12/1/0 C xy kgC/m2/s C2H6 - 1 100 0 C2H6_2010_biofuel $ROOT/C2H6_2010/v2017-05/C2H6_global_anth_biof.2010$MM.nc BIOFUEL_C2H6 2010/1-12/1/0 C xy kgC/m2/s C2H6 - 2 100 )))C2H6_2010
In GEOS-Chem 12.5.0, Xiao C2H6 emissions were retired, but the recommendation was to use Xiao C3H8 emissions over CEDS C3H8 emissions to address low CEDS totals. In this version, "Xiao" was renamed to "Xiao_C3H8" to reflect that only C3H8 emissions are used and the HEMCO hierarchy was increased from a value of 5 to 10 to ensure that inventory overwrites CEDS, which has hierarchy = 5.
--> C2H6_2010 : true --> XIAO_C3H8 : true ... #============================================================================== # --- 2010 Global fossil fuel and biofuel emissions of C2H6 (Tzompa-Sosa et al. (2017) --- # # NOTES: # - These C2H6 emissions are used in place of CEDS #============================================================================== (((C2H6_2010 0 C2H6_2010_oilgas $ROOT/C2H6_2010/v2019-06/C2H6_global_anth_biof.2010$MM.4x5.nc ANTHR_C2H6 2010/1-12/1/0 C xy kgC/m2/s C2H6 - 1 100 0 C2H6_2010_biofuel $ROOT/C2H6_2010/v2019-06/C2H6_global_anth_biof.2010$MM.4x5.nc BIOFUEL_C2H6 2010/1-12/1/0 C xy kgC/m2/s C2H6 - 1 100 )))C2H6_2010 #============================================================================== # --- Xiao et al., JGR, 2008 --- # # NOTES: # - Xiao et al. C3H8 emissions are used in place of CEDS # - Priority is set to 10 to overwrite CEDS, but not newer regional inventories (NEI2011, MIX) #============================================================================== (((XIAO_C3H8 0 XIAO_C3H8 $ROOT/XIAO/v2014-09/C3H8_C2H6_ngas.geos.1x1.nc C3H8 1985/1/1/0 C xy kgC/m2/s C3H8 6/7/26/22 1 10 0 BIOFUEL_C3H8 $ROOT/BIOFUEL/v2014-07/biofuel.geos.4x5.nc BIOFUEL_C3H8 1985/1/1/0 C xy kgC/m2/s C3H8 - 1 10 )))XIAO_C3H8
As a result, C3H8 anthropogenic emissions increase globally:
###############################################################################
### Emissions totals for species C3H8 ###
### Ref = GC_12.4.0; Dev = GC_12.5.0 ###
###############################################################################
Ref Dev Dev - Ref
C3H8 Anthro : 0.399813 0.595361 0.195548 Tg C
--Melissa Sulprizio (talk) 16:38, 15 October 2019 (UTC)
Disable Br2 offline seasalt emissions to match GEOS-Chem Classic default
This update was included in GEOS-Chem 12.3.2, which was released on 02 May 2019.
There was a discrepancy between GCHP and GEOS-Chem Classic seasalt species introduced with the adoption of offline seasalt emissions in GCHP 12.0.0. The reason for the discrepancy is now identified as Br2 offline seasalt emissions being enabled in GCHP. Br2 is disabled in the GEOS-Chem online seasalt HEMCO extension and the mismatch caused differences in benchmark and standard simulation run results.
--Lizzie Lundgren (talk) 22:27, 24 April 2019 (UTC)
--Bob Yantosca (talk) 15:24, 2 May 2019 (UTC)
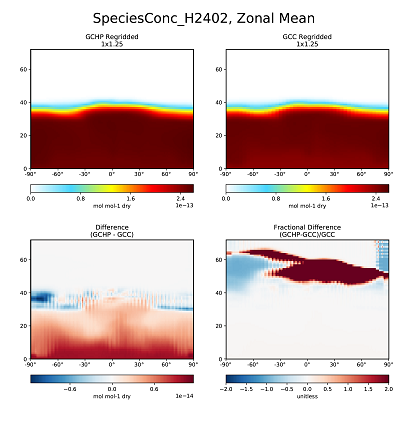
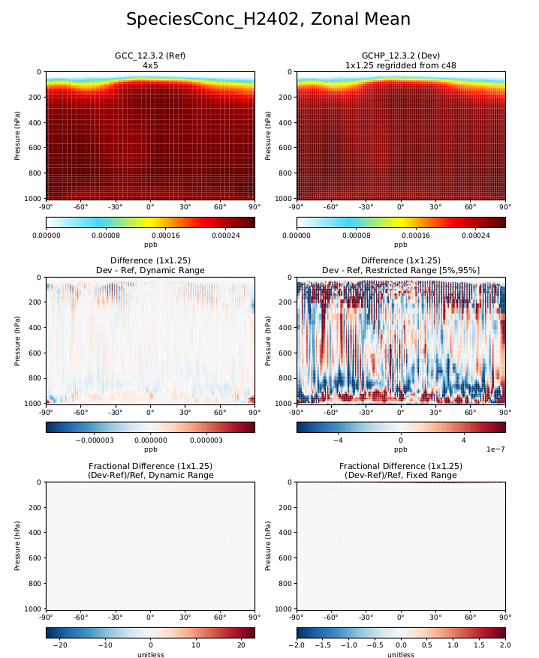
Inconsistency in H2402 concentrations in GCHP vs GEOS-Chem Classic
This update (Git ID: rel/12.3.2) was included in GEOS-Chem 12.3.2, which was released on 02 May 2019.
We have discovered an inconsistency with the concentrations of the halogen species H2402 (chemical formula: C2Br2F4) between GEOS-Chem "Classic" and GCHP. This inconsistency appears as the large fractional difference above ~50 km in the lower-right frame of the plot below.
We have traced this issue to the fact that surface boundary conditions for H2402 used by GCHP differ from those used by GEOS-Chem "Classic", and also that the boundary conditions in GEOS-Chem "Classic" are read in improperly. A fix will be made in an upcoming GEOS-Chem version.
Solution
It was discovered that this UCX input file
ExtData/CHEM_INPUTS/UCX_201403/Init1D/Montreal.nc
(which is read by GEOS-Chem "Classic" but not by GCHP) contained surface volumne mixing ratio data that was interpolated incorrectly. The file also had corrupted timestamps. This caused GEOS-Chem "Classic" to read the wrong year of data as opposed to GCHP.
Seb Eastham has created a new version of this file:
ExtData/CHEM_INPUTS/UCX_201904/Montreal_fix.nc
with the proper timestamps. Using the new file results in much better agreement between GEOS-Chem and GCHP, as shown in the plot below:
--Bob Yantosca (talk) 15:24, 2 May 2019 (UTC)
Declare State_Met%AREA_M2 as 2-dimensional, not 3-dimensional
This update (Git ID: e4af25f3b) was included in GEOS-Chem 12.3.1, which was released on 08 Apr 2019.
The State_Met%Area_M2 field (which holds grid box surface areas in m2) was allocated as (I,J,1). But this causes issues with GCHP diagnostics. When GCHP sees a 3rd dimension, it will try to save diagnostics with all levels.
The fix is to allocate State_Met%Area_M2 as an (I,J) array instead of as an (I,J,1) array.
--Bob Yantosca (talk) 19:47, 8 April 2019 (UTC)
Do not allocate memory to array MINIT in strat_chem_mod.F90
This update (Git ID: 729fd2a7) was included in GEOS-Chem 12.3.1, which was released on 08 Apr 2019.
For more information about this issue, please see this section of our Stratospheric chemistry wiki page.
--Bob Yantosca (talk) 19:47, 8 April 2019 (UTC)
Planeflight diagnostic now writes out data for the last timestep of the day
This update was be included in GEOS-Chem 12.3.1, which was released on 08 Apr 2019.
For more information about this issue, please see this section of our Planeflight diagnostic wiki page.
--Bob Yantosca (talk) 19:47, 8 April 2019 (UTC)
Tagged CO bug fixes: Fix CO units and write secondary production into a netcdf file
This update was included in GEOS-Chem 12.3.0, which was released on 01 Apr 2019.
For more information about this issue, please see this section of our Tagged CO simulation wiki page.
--Melissa Sulprizio (talk) 16:38, 28 March 2019 (UTC)
GCHP adjoint development requires time to run backwards in GCHP
This update was included in GEOS-Chem 12.3.0, which was released on 01 Apr 2019.
Limitations in MAPL previously prevented running the clock backwards in GCHP. However, running time backwards is a feature necessary to develop a GCHP adjoint model. To enable the feature, Atanas Trayanov (GMAO) provided source code updates to be incorporated into MAPL. Lizzie Lundgren implemented the update and added the feature of toggling time direction in configuration file CAP.rc at runtime.
To run GCHP backwards set REVERSE_TIME to 1. Start and end times set in runConfig.sh will then be end and start times respectively. REVERSE_TIME is set to 0 (off) by default in GCHP run directories. This feature is for use in adjoint development and should not be used for general production runs. Output using it has not been validated.
--Lizzie Lundgren (talk) 20:58, 22 March 2019 (UTC)
Further bug fix to prevent incorrect restart file output when spanning leap year days
This update was included in GEOS-Chem 12.3.0, which was released on 01 Apr 2019.
A further bug fix (in addition to the fix that was added to GEOS-Chem 12.2.1) needed to be made to the HISTORY component. This fix prevents restart files (or other instantaneous netCDF diagnostic files) from being created with data fields that are all zeroes.
It turns out that we needed to add some additional code to:
- Call routine History_SetTime from main.F before calling History_Update;
- Call routine HistContainer_UpdateIvalSet from routine History_Update for intervals longer than a month, so that the update interval would be recomputed properly if spanning a leap year day;
- Do not subtract the HeartBeat timestep in seconds from the initial update alarm. This is equivalent to advancing the clock before calling History_Update.
This fix was tested by running a simulation for Jan 1, 2016—Mar 1, 2016 (spanning a leap year day) with the Restart collection set to archive at 1 month intervals (00000100 000000). We obtained restart files with non-zero data for Feb 1st and Mar 1st. So this issue should be fixed.
--Bob Yantosca (talk) 16:32, 11 March 2019 (UTC)
Add option to always use simulation year for specified fields
This update was included in GEOS-Chem 12.3.0, which was released on 01 Apr 2019.
For more information about this issue, please see this section of our HEMCO versions wiki page.
--Melissa Sulprizio (talk) 20:18, 10 March 2019 (UTC)
Bug fix: netCDF diagnostic collections with frequency "End" now properly span leap year days
This update (Git ID: ca68f508) was included in GEOS-Chem 12.2.1, which was released on 28 Feb 2019.
Amos Tai wrote:
I have been running GEOS-Chem 12.1.1 Tropchem from 20111101 to 20111201, and that created a normal restart file for 20111201. Then I started the simulation again from 20111201 onward but now running into a problem on 20120301. After archiving the 2012 February data, GEOS-Chem created a GC restart file for 20120301 as usual, but now this restart file is empty, and obviously the simulation can’t move forward. HEMCO restart isn’t affected. I’m attaching the GC log script here, which also didn’t who any problem when archiving the restart file. I’m running it again to see if it happens again, but have you been aware of similar problems before? I suspect it may have something to do with 2012 being a leap year, so somewhere something doesn’t recognize 20120229?.
Bob Yantosca replied:
You were indeed correct, the issue was the Restart collection was not properly accounting for leap year days.
As it turns out, when I wrote the algorithm for computing "alarms" for the Update, File Write, and File Close operations, I had assumed that users would only request diagnostic output at intervals of less than 1 month, exactly 1 month, or exactly 1 year. These are the most common diagnostic intervals and these are the easiest to code up. So technically this wasn’t a "bug" per se, but rather a "design limitation" (which is just as bad sometimes).
When we updated the 12.1.0 code to define the restart files as a netCDF collection with frequency "End", then the existing algorithm would would fail if the interval spanned a leap year day. I have since updated the algorithm for computing "alarms" for the Update, File Write, and File Close operations so that they now work properly when spanning leap years. If you start a run on a leap year such as 2016/01/01 and run to 2016/03/01, then you will now get a restart file for 2016/03/01. I also tested the case if you start e.g. on 2015/12/01 and then run past New Year's Day to 2016/04/01, you will get output on 2016/04/01, as expected. So this should work OK now.
NOTE: There may still be some cases where the new algorithm will fail. Using the frequency "End" should work well for intervals less than a year. If you are submitting individual jobs to a batch scheduler, and the jobs are only a couple of months long, then it should be OK to use "End" for the Restart collection. But if you are jobs of 1 year or longer to the scheduler, we would recommend that you set the Restart collection frequency to either every month or every other month. This will be a failsafe in case the run dies for any reason...you would only have to rerun from the last restart instead of having to rerun the last year.
--Bob Yantosca (talk) 16:29, 27 February 2019 (UTC)
Use online sea flux emissions by default in GCHP
This update (Git ID: 9759d26b) was included in GEOS-Chem 12.2.1, which was released on 28 Feb 2019.
Offline sea flux emissions generated with a 1-year GEOS-Chem Classic simulation at 2x2.5 are used in GCHP by default since 12.0.0. However, the offline emissions cause greater differences between GEOS-Chem Classic and GCHP than online sea flux emissions. In order to make GEOS-Chem Classic and GCHP more comparable in benchmark simulations the default will be to use online sea flux beginning in version 12.2.1. Investigation of the offline sea flux emissions inventory is needed before making the files default.
--Lizzie Lundgren (talk) 19:18, 26 February 2019 (UTC)
Missing scaling factors for MERRA-2 simulations with GCHP
This update was included in GEOS-Chem 12.2.1, which was released on 28 Feb 2019.
Seb Eastham wrote:
- I noticed that there’s an issue with the ExtData.rc template in the GCHP/Run directory. The pressure fields (SLP, PS, and TROPPT) are all expected to be in hPa. This is true for GEOS-FP, where those fields were indeed stored in hPa, but the MERRA-2 pressure data is stored in Pa. As a result, any GCHP simulations using MERRA-2 will have weird behavior.
- This is an easy fix, fortunately. Whenever a MERRA-2 simulation dir is generated, the scaling factor (the second “none” on each line for SLP/PS/TROPPT) needs to be changed to 0.01. That should fix the issue. Might be worth putting some sanity checks in e.g. Includes_Before_Run.H which throw a warning (not necessarily an error – always possible someone will one day want to simulate Venus or something with GEOS-Chem) if any pressure values are > 5000 (ie a value way outside the bounds of normality but low enough that it will be triggered it we have an order-of-magnitude error due to units). Could be worth also checking if any values of TROPPT, SLP, PS1, or PS2 are < e.g. 200, for the same reason (e.g. someone uses GEOS-FP but thinks the units are Pa).
--Lizzie Lundgren (talk) 17:55, 19 February 2019 (UTC)
GCHP uses outdated MODIS files missing projection error fix
This update was included in GEOS-Chem 12.2.1, which was released on 28 Feb 2019.
The MODIS LAI files used in GEOS-Chem Classic were updated in v11-02f to address an issue of offsetting along the southern coasts of Australia. Some SW ocean grid boxes had LAI > 0 and some SE land boxes had LAI = 0. However, the source directory was not updated in GCHP and the older files were used in GCHP versions v11-02f through 12.2.0.
To correct this issue the GCHP run directory will be updated to use the newer files in 12.2.1. For more information on the MODIS LAI update, please see the wiki post for Global 0.25x0.25 MEGAN input and MODIS LAI.
--Lizzie Lundgren (talk) 17:51, 19 February 2019 (UTC)
Fix Henry's Law coefficient for C2H6
This update was included in GEOS-Chem 12.2.1, which was released on 28 Feb 2019.
Chris Holmes wrote:
- I’ve attached a bugfix for the Henry’s Law coefficient for C2H6. The exponent sign was wrong, but there seems to be no effect on GC results because C2H6 does not have dry or wet deposition.
In Headers/species_database_mod.F90, change the following line:
#if defined( NEW_HENRY_CONSTANTS ) Henry_K0 = 1.90e+5_f8 * To_M_atm Henry_K0 = 1.90e-5_f8 * To_M_atm Henry_CR = 2400.0_f8 #endif
--Melissa Sulprizio (talk) 16:01, 8 February 2019 (UTC)
--Bob Yantosca (talk) 18:55, 28 February 2019 (UTC)
ESMF version update required for MAPL upgrade
This update was included in GEOS-Chem 12.2.0, which was released on 19 Feb 2019.
GCHP versions 12.1.1 and prior use ESMF v5.2.0rp2 which was released in 2012. Besides this being a very old version that warrants updating, an update to a more recent version of ESMF is required for compatibility with new functionalities GMAO is incorporating into MAPL. Prior to implementing the MAPL update into GCHP we therefore must update ESMF and plan to use the last public release of ESMF, v7.1.0r. For more information see the ESMF documentation at www.earthsystemcog.org/projects/esmf.
--Lizzie Lundgren (talk) 18:36, 17 January 2019 (UTC)
Fix bug in stratosperic aerosol surface area netcdf diagnostics
This update (Git ID: e4fecf01) was included in GEOS-Chem 12.2.0, which was released on 19 Feb 2019.
This update was submitted by Jonathan Moch who pointed out that "As written, the code is filling the stratospheric liquid aerosol surface area density and the polar strat cloud surface area with the stratospheric liquid aerosol surface area and then refilling the values with polar strat surface area. So if you aren't running the model inside GEOS-5 the AerSurfAreaPolarStratCloud and AerSurfAreaStratLiquid are identical, which shouldn't be the case."
A fix for this was implemented by Christoph Keller in the version of GEOS-Chem used in GEOS-5. To fix the issue in the offline model we use the same solution.
--Lizzie Lundgren (talk) 23:30, 4 January 2019 (UTC)
GCHP Bug fix: use checkpoint output file as subsequent run restart
This fix was included in GEOS-Chem 12.2.0, which was released on 19 Feb 2019.
This bug fix applies only to the multi-run option of GCHP. It corrects a previous silent bug in which the initial restart file specified in runConfig.sh was used as the restart when doing multiple subsequent runs (i.e. in the presence of cap_restart). The proper behavior when using the multirun option (runScriptSamples/multirun.sh) is to use the checkpoint file (snapshot of end of last run) if the run is continuing from where the last run left off.
This is now corrected by automatically updating the entry for the initial restart file in GCHP.rc with the name of the checkpoint file if the file cap_restart is present just prior to execution. For simplicity the checkpoint file no longer contains the grid resolution in its name.
--Lizzie Lundgren (talk) 20:53, 4 January 2019 (UTC)
GCHP transport fix to prevent instant propagation from troposphere to stratosphere
This fix was included in 12.1.1, which was released on 13 Dec 2018 .
Sebastian Eastham describes this bug fix as follows:
GCHP uses a simple mass conservation kludge which rescales global tracer concentrations between transport steps. This is equivalent to the pressure fixer in GC-Classic, used to account for inconsistency between the surface pressure field (which sets the vertical grid spacing up to the upper troposphere) and the averaged wind speeds. Although the scaling factor is usually small, this can cause nonphysical redistribution of mass. This is especially problematic because mass can be transferred to, or from, the stratosphere, which should be impossible given that the stratosphere operates on fixed pressure levels. In a future update, this problem will be resolved by direct application of mass fluxes archived from GEOS in place of winds, but while that update is in development we have produced an interim fix
This patch forces mass redistribution to be constrained to the variable pressure levels in the troposphere. Although this slightly increases spurious mixing in the troposphere, it prevents changes in the troposphere from being instantly propagated to the stratosphere. This can have significant effects when inspecting (e.g.) the response to changes in surface emissions of NOx.
--Lizzie Lundgren (talk) 16:03, 6 December 2018 (UTC)
--Bob Yantosca (talk) 15:50, 13 December 2018 (UTC)
Always enable BCPH_TPBC to allow saving out BC files in global simulations
This update (Git ID: 072ec9d6) was included in 12.1.1, which was released on 13 Dec 2018 .
Yanxu Zhang reported an issue in GEOS-Chem 12.1.0 where BC files would not save out, even when turned on in the Nested Grid Menu of input.geos. To resolve this issue, we will always add USER_DEFS += -DBPCH_TPBC in Makefile_header.mk. The switch to turn on saving out boundary condition files is still controlled by the options in input.geos, but this fix ensures the tpcore_bc_mod.F code is always compiled.
--Melissa Sulprizio (talk) 15:40, 30 November 2018 (UTC)
--Bob Yantosca (talk) 15:49, 13 December 2018 (UTC)
Skip planeflight observations outside a nested domain
This update (Git ID: f6f90fd4) was included in 12.1.1, which was released on 13 Dec 2018 .
Lei Zhu provided a fix for planeflight_mod.F to skip flight observations outside the nested domains. With the fix, the code works for all 0.5 and 0.25 nested domains. For more information please see the pull request on Github.
--Melissa Sulprizio (talk) 13:29, 27 November 2018 (UTC)
--Bob Yantosca (talk) 15:48, 13 December 2018 (UTC)
Allow for SZA to 98 degrees when getting photolysis rates
This update was included in GEOS-Chem 12.1.0, which was released on 26 Nov 2018.
Eric Palmer (GMAO) wrote:
I see that the code that calls the photolysis rates (in flexchem_mod.F90), is only called when the solar zenith angle is < 90 degrees (COS(SZA) > 0.). However, Fast-Jx allows for SZA down to 98 degrees, which is important in the stratosphere-mesosphere where sunlight still illuminates at high altitudes if the sun is below the horizon at the surface. To allow for SZA down to 98 deg, I made the following one line change:
IF ( State_Met%SUNCOSmid(I,J) > 0.e+0_fp ) THEN
to:
IF ( State_Met%SUNCOSmid(I,J) > -0.13917e+0_fp ) THEN
where -0.13917e+0_fp = COS(98 degrees) This looks like it has the desired effect, as species with sharp gradients across the terminator (eg: Br, BrO), are now more "rounded" and look much more like GMI (which also allows SZA down to 98 degrees). This also eliminates the problem with the non-conservation of bromine family species in the mesosphere due to transport across the terminator (ie, the sharp edges of a square wave).
--Lizzie Lundgren (talk) 14:59, 16 October 2018 (UTC)
--Bob Yantosca (talk) 16:09, 26 November 2018 (UTC)
Fix bug in application of CFC surface mixing ratios in GCHP
This update was included in GEOS-Chem 12.0.3, which was released on 16 Oct 2018.
This fix is urgent, as any current users of the last several versions of GCHP will be see bad results for relevant species. We recommend that all GCHP users implement this update if not updating to the latest version.
Sebastian Eastham describes this bug as follows:
Basically, we specify a target year for CFC emissions to be read in in input.geos, so (eg) we can specify “2050” and all surface VMRs for CFCs will be set according to the expected value for 2050. Usually, though, we use a value of 0 – meaning “use the current year”. In GCC, the value Input_Opt%CFCYEAR is set to Get_Year () during initialization, and the offset is calculated in UCX_mod, meaning the right year gets used. However, in GCHP this isn’t possible because Get_Year doesn’t return the right value until after initialization. There was a workaround in UCX_mod already where we had a local variable “CFCYEAR” which was a copy of Input_Opt%CFCYEAR, and which got set to Get_Year() at runtime if it was still zero; however, this wasn’t being propagated in the case of an ESMF environment (where UCXNETCDF is .False.) because we only passed Input_Opt rather than the updated local variable. As a result, the CFC “emissions” were always for 1950 rather than for the true target year.
--Lizzie Lundgren (talk) 21:18, 15 October 2018 (UTC)
--Bob Yantosca (talk) 16:10, 26 November 2018 (UTC)
Fix incorrect run configuration for offline sea salt emissions in GCHP
This update was included in GEOS-Chem 12.0.3, which was released on 16 Oct 2018.
The offline sea salt emissions applied in GCHP starting in 12.0.0 contained a bug due to incorrect settings in HEMCO_Config.rc. SALA and SALC sea salt values read from file should be scaled and applied to BrSALA and BrSALC respectably. However, the HEMCO_Config.rc settings resulted in applying the scaled values to SALA and SALC instead. This bug was identified by Seb Eastham (MIT) and is corrected by updating the species column in HEMCO_Config.rc. No source code updates are necessary but you must create a new run directory or apply the fix in your existing run directories.
--Lizzie Lundgren (talk) 21:13, 15 October 2018 (UTC)
--Bob Yantosca (talk) 16:10, 26 November 2018 (UTC)
Update HEMCO configuration file for tagged CO simulation
This update (Git ID: 02aaad73) was included in GEOS-Chem 12.1.0, which was released on 26 Nov 2018.
The tagged CO simulation was still using NEI2005 emissions over the US by default, while all other simulations had retired the NEI2005 option and were using NEI2011 instead. For consistency, we will use NEI2011 with the tagged CO simulation and remove NEI2005.
In addition, the CEDS and DICE_Africa inventories were omitted from tagged CO HEMCO configuration file. To maintain consistency with the full-chemistry simulations, these inventories are now added for tagged CO and are turned on by default.
--Melissa Sulprizio (talk) 17:37, 21 September 2018 (UTC)
--Bob Yantosca (talk) 16:10, 26 November 2018 (UTC)
Fix regional tile file used for APEI inventory in GCHP
This bug fix only requires downloading updated files from gcgrid/ExtData/GCHP/TileFiles
The regional tile file used by GCHP for regrigding the APEI inventory from native resolution to cubed sphere was incorrect at the time of the 12.0.0 release. The tile file erroneously caused the APEI inventory to be regridded at a 180 degree longitude offset. Corrected tile files now replace the erroneous ones at gcgrid/ExtData/GCHP/TileFiles. No changes are necessary to the model source code or the GCHP run directory.
--Lizzie Lundgren (talk) 17:30, 19 September 2018 (UTC)
Fix masked anthropogenic emissions bug in GCHP
This update was included in GEOS-Chem 12.1.0, which was released on 26 Nov 2018.
Integration of the DICE-Africa inventory in GCHP resulted in erroneously masking out total global anthropogenic emissions outside of Africa. Updating the Africa mask file from containing regional bounds to global bounds corrects the problem. GCHP users who are using the DICE-inventory prior to 12.1.0 can fix the issue by updating their Africa mask file to MASKS/v2014-07/AF_LANDMASK.geos.05x0666.global.nc which is now available on gcgrid.
--Lizzie Lundgren (talk) 16:50, 14 September 2018 (UTC)
--Bob Yantosca (talk) 16:11, 26 November 2018 (UTC)
Add support for continuous integration with TravisCI
This update was included in GEOS-Chem 12.1.0, which was released on 26 Nov 2018.
We have now linked the GEOS-Chem Github repository to the continuous integration tool TravisCI. Each time a commit is pushed to the repository, TravisCI will perform a test to ensure that the commit will not "break" GEOS_Chem. Right now, TravisCI only compiles GEOS-Chem, but we will eventually implement compile-and-run tests in the near future.
You can see the TravisCI status page for GEOS-Chem here: https://travis-ci.com/geoschem/geos-chem/builds
Also, if you navigate to the "commits" page of the GEOS-Chem Github repository, you will see a green checkmark next to each commit that was checked by TravisCI:
NOTE: As of this writing, only the dev/12.1.0 branch is being checked by TravisCI. When we merge this into the master branch, then all future branches will be checked.
--Bob Yantosca (talk) 16:11, 26 November 2018 (UTC)
Fixed local-time averaging in ND51 and ND51b satellite timeseries diagnostics
This update (Git ID: 14b29d3d) was included in GEOS-Chem 12.0.2, which was released on 10 Oct 2018.
Bo Zhang wrote:
We encountered a potential issue in the satellite diagnostic ND51b in GEOS-Chem v11-01. We find some kind of discontinuity in total column AOD (a sum of all aerosol species) along the west-east direction. It feels like data are missing/reduced occasionally in every other pixels alone the W-E direction. We are running v11-01 with full chemistry + UCX driven by MERRA2. The same problem remains after switching to MERRA. We checked AOD maps in monthly output which looks normal.
Bob Yantosca replied:
In recent versions of GEOS-Chem, the number of advected species increased (to accommodate additional halogen and SOA species). So we had to increase the non-species tracer numbers to 500:
! ND51 tracer numbers:
! ============================================================================
! 1 - nAdvect : GEOS-CHEM advected species [v/v ]
! 501 : OH concentration [molec/cm3 ]
! 502 : NOy concentration [v/v ]
! 503 : Relative Humidity [% ]
! ... etc ...
Complicating matters further, some ND51/ND51b quantities, including the AOD’s, were only updated on every chemistry timestep (20 mins) instead of on every dynamic timestep (10 mins). There was a CASE statement in routine WRITE_DIAG51 to divide by the proper counting array to create the average quantity:
SELECT CASE( Input_Opt%ND51_TRACERS(W) )
CASE( 91, 92, 76, 77 )
... etc ...
CASE( 84:89, 115:121 )
... etc ...
CASE( 105:114 )
... etc ...
CASE DEFAULT
... etc ...
END SELECT
But we just realized that this CASE statement had the old ND51/ND51b tracer numbers ( < 500) and not the new ones.
Therefore, we have made the following fixes, which should eliminate any striping or discontinuities
- Archive most ND51/ND51b tracers (except for NOy, which has special handling) on every dynamic timestep. Remove the GOOD_CHEM, GOOD_CT_CHEM, GOOD_EMIS, GOOD_CT_EMIS arrays.
- Update the CASE statement shown above so that all tracers (except NOy) are divided by the number of dynamic timesteps when each grid box was within the proper local time range.
Also note: the ND51/ND51b diagnostics were developed in an ad-hoc fashion and were were never really thought out holistically. It is possible that there may be some issues with some of the other quantities in ND51/ND51b, but the GCST is not planning on investigating further. The bpch diagnostics will be eventually removed from GEOS-Chem once we have a satisfactory equivalent local-time averaging netCDF diagnostic.
--Bob Yantosca (talk) 15:12, 10 October 2018 (UTC)
Fix typo when registering the State_Chm%SSAlk field
This update (Git ID: 55d16873) was included in GEOS-Chem 12.0.1, which was released on 24 Aug 2018.
Shuting Zhai wrote:
I found a typo in state_chm_mod.F90 where we register the State_Chm%SSAlk field. I believe this line:
CALL Register_ChmField( ... State_Chm%pHCloud, ... )
Should be:
CALL Register_ChmField( ... State_Chm%SSAlk, ... )
Upon fixing this issue, the GCST discovered that the State_Chm%SSAlk array cannot be registered as a single array because it is dimensioned as (IIPAR,JJPAR,LLPAR,2). When registering quantities, the 4th dimension is always interpreted as the number of chemical species. Therefore, registering State_Chm%SSAlk in this way caused an error.
The solution is to register the two dimensions of State_Chm%SSAlk separately:
- SSAlkAccum points to State_Chm%SSAlk(:,:,:,1)
- SSAlkCoarse points to State_Chm%SSAlk(:,:,:,2)
This update will require you to list Chem_SSAlkAccum and Chem_SSAlkCoarse as requested diagnostics instead of the single field Chem_SSAlk in the HISTORY.rc file ( which specifies input options for netCDF diagnostics).
--Bob Yantosca (talk) 15:33, 23 August 2018 (UTC)
Fixes for compilation issues
We have added several fixes for issues encountered during GEOS-Chem compilation:
- Add a more robust test for netCDF-Fortran in Makefile_header.mk
- Fixes for compilation issues encountered with GNU Fortran 8.2.0
- Fixes for compilation issues encountered with Intel Fortran 18
--Bob Yantosca (talk) 14:26, 22 August 2018 (UTC)
Fix mis-indexing in the ND05 diagnostic
This update was included in GEOS-Chem 12.0.1, which was released on 24 Aug 2018.
For a complete description of this issue, please see this post on our Sulfate aerosols wiki page.
--Bob Yantosca (talk) 14:18, 22 August 2018 (UTC)
Bug fix for HEMCO soil NOx error with ifort 17
This fix was included in GEOS-Chem 12.0.0.
For complete information about this issue and how it was resolved, please see this post on our Implementation of HEMCO in GEOS-Chem wiki page..
--Bob Yantosca (talk) 13:54, 27 June 2018 (UTC)
FlexChem bug fix: do not zero ACTA, EOH, HCOOH
This fix was included in GEOS-Chem 12.0.0.
For complete information about this bug and how it was fixed, please see this post on our FlexChem wiki page.
--Bob Yantosca (talk) 14:42, 27 June 2018 (UTC)
Avoid double counting CEDS and GEIA NH3 biofuel emissions
This fix was included in GEOS-Chem 12.0.0.
Eloise Marais wrote:
If CEDS includes NH3 emissions, then this will be double counting. I recommend using CEDS, as GEIA is very out of date (1998 according to the HEMCO configuration file I have).
Daniel Jacob replied:
We should use the GEIA NH3 natural emission inventory (1° x 1° file from Eleanor Morris) but not the GEIA NH3 biofuel inventory.
UPDATE 29 Jun 2018:
We discovered that HEMCO's default behavior is to ignore the GEIA NH3 anthro and biofuel emissions if CEDS is turned on. This is shown in the HEMCO.log file when VERBOSE = 3:
Register_Base: Checking GEIA_NH3_ANTH HEMCO: Entering Get_targetID (hco_config_mod.F90) ( 4) Skip container GEIA_NH3_ANTH because of CEDS_NH3_WST . . . Register_Base: Checking GEIA_NH3_BIOF HEMCO: Entering Get_targetID (hco_config_mod.F90) ( 4) Skip container GEIA_NH3_BIOF because of CEDS_NH3_WST_Cat00002
But for the sake of clarity, we will modify the HEMCO_Config.rc files to explicitly request this behavior:
(((.not.CEDS.or.EDGARv43 # GEIA NH3 anthro (skip if CEDS or EDGARv43 is on) 0 GEIA_NH3_ANTH $ROOT/NH3/v2014-07/NH3_geos.4x5.nc NH3_ANTH 1990/1-12/1/0 C xy kg/m2/s NH3 - 1 1 # GEIA NH3 biofuel (skip if CEDS or EDGARv43 is on) (((BIOFUEL 0 GEIA_NH3_BIOF $ROOT/NH3/v2014-07/NH3_biofuel.geos.1x1.nc NH3_BIOFUEL 1998/1-12/1/0 C xy kg/m2/s NH3 - 2 1 )))BIOFUEL ))).not.CEDS.or.EDGARv43
--Bob Yantosca (talk) 21:02, 29 June 2018 (UTC)
Fix bug in ARCTAS_SHIP_SO2 scale factors
This fix was included in GEOS-Chem 12.0.0.
For a complete description of this issue and how it was resolved, please see this post on our Ship Emissions wiki page.
--Bob Yantosca (talk) 15:54, 3 July 2018 (UTC)
Fix issues in complexSOA and complexSOA_SVPOA restart files
This fix was included in GEOS-Chem 12.0.0.
Eloise Marais wrote:
I just wanted to draw your attention to a few issues with the restart files provided on the ftp site (http://ftp.as.harvard.edu/gcgrid/data/ExtData/SPC_RESTARTS/). The complexSOA and complexSOA_SVPOA restart files (4° x 5° and 2° x 2.5°) both have a species entry for IEPOX and also IEPOXD, IEPOXB, and IEPOXA, but should only have the latter 3. This is the same for RIP. The number of species in the 4° x 5° and 2° x 2.5° files for complexSOA is also different.
To fix these issues, we have generated new restart files for the complexSOA and complexSOA_SVPOA simulations. You can download these to your SPC_RESTARTS folder with the following commands:
wget "ftp://ftp.as.harvard.edu/gcgrid/data/ExtData/SPC_RESTARTS/initial_GEOSChem_rst.2x25_complexSOA.nc" wget "ftp://ftp.as.harvard.edu/gcgrid/data/ExtData/SPC_RESTARTS/initial_GEOSChem_rst.2x25_complexSOA_SVPOA.nc" wget "ftp://ftp.as.harvard.edu/gcgrid/data/ExtData/SPC_RESTARTS/initial_GEOSChem_rst.4x5_complexSOA.nc" wget "ftp://ftp.as.harvard.edu/gcgrid/data/ExtData/SPC_RESTARTS/initial_GEOSChem_rst.4x5_complexSOA_SVPOA.nc"
--Bob Yantosca (talk) 18:01, 5 July 2018 (UTC)
Bug fix for reference time in netCDF diagnostic files
This fix was included in GEOS-Chem 12.0.0.
Maria Zatko wrote:
For the sake of run time, we would like to run GEOS-Chem with the old (15/30 minute timesteps) instead of the recommended timesteps (10/20 minutes). I noticed that the output netcdf files have different timestamps depending on the timestep (e.g., 10/20 minutes, 15/30 minutes) used. For example, when 10/20 minute timesteps are used, the time units in the following netcdf output files range from 0 to 23 UTC:
GEOSChem3hrInst.SpeciesConcWithMet.20150701_0000z.nc4 (file size: 4.6 GB)
GEOSChem3hrInst.SpeciesConcWithMet.20150702_0000z.nc4 (file size: 5.3 GB)
time:units = "minutes since 2015-07-01 00:00:00 UTC"
When 15/30 minute timesteps are used, the time units in the following output files include hour 24:
GEOSChem3hrInst.SpeciesConcWithMet.20150701_0000z.nc4 (file size: 4.6 GB)
GEOSChem3hrInst.SpeciesConcWithMet.20150701_2400z.nc4 (file size: 5.3 GB)
GEOSChem3hrInst.SpeciesConcWithMet.20150702_2400z.nc4 (file size: 5.3 GB)
time:units = "minutes since 2015-07-01 24:00:00 UTC"
I wanted to check in to see if others have run into this issue as well.
Bob Yantosca replied:
This issue is caused by floating-point roundoff error in converting from the Julian date to the calendar date. I have created a more robust fix for this issue, which now computes the correct netCDF file reference time independent of the model "heartbeat" timestep.
To apply the fix:
- Copy the patch file ftp://ftp.as.harvard.edu/pub/exchange/bmy/nc_ref_time_fix.txt to your computational server.
- Change to your GEOS-Chem v11-02 release candidate code directory and check out the "master" branch.
- Type: git am < nc_ref_time_fix.txt
The fix will be standardized in the GEOS-Chem 12.0.0 release.
--Bob Yantosca (talk) 15:10, 12 July 2018 (UTC)
Bug fix for CH4 boundary conditions
This fix was included in GEOS-Chem 12.0.0.
Lee Murray wrote:
I’ve been testing the release candidate, and believe that we need to change the default input.geos options for the Standard and Benchmark simulations in the unit tester to :
%%% EMISSIONS MENU %%% :
Turn on emissions? : T
HEMCO Input file : HEMCO_Config.rc
=> 1ppt MBL BRO Sim.? : F
Switches for UCX :---
=> Use CH4 emissions? : F
=> Turn on surface BCs :---
=> CH4? : F
=> OCS? : T
=> CFCs? : T
=> Cl species? : T
=> Br species? : F
=> N2O? : T
=> Set initial glob MRs:---
=> strat. H2O? : T
=> CFC emission year : 0
By default in the unit tester, => CH4 is still enabled. This appears to set up conflict between the older UCX surface layer and the newer HEMCO methane PBL boundary conditions, causing air to be a mix of preindustrial and present-day values (see below for Jul 1, 2013 surface methane in a benchmark run). Setting it to F and/or removing LCH4SBC causes the methane to behave as expected, i.e., being set by HEMCO in all simulations, with the expected spatial pattern. Also, we can slate Online CH4 chemistry? : T in the Chemistry Menu and Input_Opt%LCH4CHEM for future removal, as it no longer does anything.
--Melissa Sulprizio (talk) 18:16, 16 July 2018 (UTC)
Fixed typo in call to GAMMA_HOBr_AER
This fix was included in GEOS-Chem 12.0.0.
Xuan Wang wrote:
When looking back at the standard GC codes of v11-02d, I found a bug in the function HETHOBr_SS_JS (in module KPP/Standard/gckpp_HetRates.F90). When calling the gamma calculation, in v11-02d, around line 3514, we have:
CALL Gamma_HOBr_AER(rAer, denAir, 2, TK, clConc, brConc, &
hConc, GAM_HOBr, r_gp)
This will cause both HOBr+Cl- (X=1) and HOBr+Br- (X=2) on sea salt use the gamma of HOBr + Br- (X=2). It should be:
CALL Gamma_HOBr_AER(rAer, denAir, X, TK, clConc, brConc, &
hConc, GAM_HOBr, r_gp)
It looks like a typo which could lead to quite different result for halogen chemistry.
--Bob Yantosca (talk) 20:26, 17 July 2018 (UTC)
Bug fix for liquid water content in gckpp_HetRates.F90
This fix was included in GEOS-Chem 12.0.0.
Jiayue Huang wrote:
Qianjie Chen and I think that there is potential a bug in KPP/Standard/gckpp_HetRates.F90 for halogen chemistry in the model v11-02d/e/f.
In the original code, we were using the grid-box average liquid water content, which likely underestimates the in-cloud Br- and Cl- concentration. So we think that we should change it to in-cloud liquid water content by dividing the cloud fraction. This results in increases the S(IV)+Br reaction in cloud, and thus reduces the BrO abundance, mainly in the 30-90N region.
This bug fix is to use cloud fraction when calculating liquid water content [cm3 H2O/cm3 air] in routine GET_HALIDE_CLDConc, which is used to calculate the in-cloud concentrations of Br- and Cl-.
--Bob Yantosca (talk) 15:48, 20 July 2018 (UTC)
Add PH2O2 to the default list of prod/loss families in the KPP mechanisms
This fix was included in GEOS-Chem 12.0.0.
The P(H2O2) field is required as input in the aerosol-only simulations. To simplify saving out new oxidant fields, we now include P(H2O2) by default in the prod/loss families so that it is saved out when users turn on the prod/loss diagnostic in either bpch or netCDF format.
--Bob Yantosca (talk) 16:16, 20 July 2018 (UTC)
Fixed typo that prevented PM2.5 from being archived to netCDF output
This fix was included in GEOS-Chem 12.0.0.
In routine Set_AerMass_Diagnostic of module GeosCore/diagnostics_mod.F90, there was a typo in the IF block where the PM2.5 diagnostic values get copied into State_Diag%PM25. The code in RED below:
IF ( Archive_BetaNO ) THEN
State_Diag%PM25(I,J,L) = ...etc...
needs to be replaced by the code in GREEN:
IF ( Archive_PM25 ) THEN
State_Diag%PM25(I,J,L) = ...etc...
--Bob Yantosca (talk) 20:33, 24 July 2018 (UTC)
Add entries to HEMCO_Config.rc for SOAP and pFe in CEDS inventory
This fix was included in GEOS-Chem 12.0.0.
The CEDS emissions entries in HEMCO_Config.rc were missing for species SOAP and pFe. To fix this, add the following lines in GREEN:
0 CEDS_CO_AGR $ROOT/CEDS/v2018-04/CO-em-anthro_CMIP_CEDS_195001-201412.nc CO_agr 1950-2014/1-12/1/0 C xy kg/m2/s CO 26 1/2/12 5 0 CEDS_SOAP_AGR - - - - - - SOAP 26/280 1/2/12 5 0 CEDS_CO_ENE $ROOT/CEDS/v2018-04/CO-em-anthro_CMIP_CEDS_195001-201412.nc CO_ene 1950-2014/1-12/1/0 C xy kg/m2/s CO 26 1/2/12 5 0 CEDS_SOAP_ENE - - - - - - SOAP 26/280 1/2/12 5 0 CEDS_CO_IND $ROOT/CEDS/v2018-04/CO-em-anthro_CMIP_CEDS_195001-201412.nc CO_ind 1950-2014/1-12/1/0 C xy kg/m2/s CO 26 1/2/12 5 0 CEDS_SOAP_IND - - - - - - SOAP 26/280 1/2/12 5 0 CEDS_CO_TRA $ROOT/CEDS/v2018-04/CO-em-anthro_CMIP_CEDS_195001-201412.nc CO_tra 1950-2014/1-12/1/0 C xy kg/m2/s CO 26 1/2/12 5 0 CEDS_SOAP_TRA - - - - - - SOAP 26/280 1/2/12 5 0 CEDS_CO_SLV $ROOT/CEDS/v2018-04/CO-em-anthro_CMIP_CEDS_195001-201412.nc CO_slv 1950-2014/1-12/1/0 C xy kg/m2/s CO 26 1/2/12 5 0 CEDS_SOAP_SLV - - - - - - SOAP 26/280 1/2/12 5 0 CEDS_CO_WST $ROOT/CEDS/v2018-04/CO-em-anthro_CMIP_CEDS_195001-201412.nc CO_wst 1950-2014/1-12/1/0 C xy kg/m2/s CO 26 1/2/12 5 0 CEDS_SOAP_WST - - - - - - SOAP 26/280 1/2/12 5 0 CEDS_SO2_AGR $ROOT/CEDS/v2018-04/SO2-em-anthro_CMIP_CEDS_195001-201412.nc SO2_agr 1950-2014/1-12/1/0 C xy kg/m2/s SO2 - 1/2/12 5 0 CEDS_SO4_AGR - - - - - - SO4 63 1/2/12 5 0 CEDS_pFe_AGR - - - - - - pFe 66 1/2/12 5 0 CEDS_SO2_ENE $ROOT/CEDS/v2018-04/SO2-em-anthro_CMIP_CEDS_195001-201412.nc SO2_ene 1950-2014/1-12/1/0 C xy kg/m2/s SO2 - 1/2/12 5 0 CEDS_SO4_ENE - - - - - - SO4 63 1/2/12 5 0 CEDS_pFe_ENE - - - - - - pFe 66 1/2/12 5 0 CEDS_SO2_IND $ROOT/CEDS/v2018-04/SO2-em-anthro_CMIP_CEDS_195001-201412.nc SO2_ind 1950-2014/1-12/1/0 C xy kg/m2/s SO2 - 1/2/12 5 0 CEDS_SO4_IND - - - - - - SO4 63 1/2/12 5 0 CEDS_pFe_IND - - - - - - pFe 66 1/2/12 5 0 CEDS_SO2_TRA $ROOT/CEDS/v2018-04/SO2-em-anthro_CMIP_CEDS_195001-201412.nc SO2_tra 1950-2014/1-12/1/0 C xy kg/m2/s SO2 - 1/2/12 5 0 CEDS_SO4_TRA - - - - - - SO4 63 1/2/12 5 0 CEDS_pFe_TRA - - - - - - pFe 66 1/2/12 5 0 CEDS_SO2_SLV $ROOT/CEDS/v2018-04/SO2-em-anthro_CMIP_CEDS_195001-201412.nc SO2_slv 1950-2014/1-12/1/0 C xy kg/m2/s SO2 - 1/2/12 5 0 CEDS_SO4_SLV - - - - - - SO4 63 1/2/12 5 0 CEDS_pFe_SLV - - - - - - pFe 66 1/2/12 5 0 CEDS_SO2_WST $ROOT/CEDS/v2018-04/SO2-em-anthro_CMIP_CEDS_195001-201412.nc SO2_wst 1950-2014/1-12/1/0 C xy kg/m2/s SO2 - 1/2/12 5 0 CEDS_SO4_WST - - - - - - SO4 63 1/2/12 5 0 CEDS_pFe_WST - - - - - - pFe 66 1/2/12 5
--Melissa Sulprizio (talk) 16:07, 27 July 2018 (UTC)
Remove duplicate C3HI spectrum in FJX_spec.dat and FJX_j2j.dat
This fix was included in GEOS-Chem 12.0.0.
Tomas Sherwen wrote:
- I've just found that both FJX_spec.dat the redundant CH3I spectrum and the new spectrum are both present in FJX_spec.dat and FJX_j2j.dat (in v12.0.0). This will have been due to the spectrum in my branch being under the species alias of "CH3IT" to preserve (and ignore) the use of "CH3I" in the namespace of the code for the existing methyl iodide simulation when the iodine simulation was developed. The "CH3IT" variables were replaced "CH3I" in the v11-1-Patches branches with halogen chemistry provided to GCST, but the existing methyl iodide simulation variable was not removed. There are no double ups in the KPP mechanism itself.
- Please remove lines 276-281 in FJX_spec.dat (copied below).
"
CH3I 243 0.000E+00 0.000E+00 0.000E+00 0.000E+00 0.000E+00 0.000E+00 J10
3.958E-20 4.769E-20 8.257E-19 2.154E-19 7.349E-20 3.351E-20
1.034E-20 4.435E-21 2.272E-21 4.393E-22 4.052E-24 0.000E+00
CH3I 300 0.000E+00 0.000E+00 0.000E+00 0.000E+00 0.000E+00 0.000E+00 J10
3.958E-20 4.769E-20 8.376E-19 2.438E-19 9.355E-20 4.680E-20
1.529E-20 6.532E-21 3.328E-21 6.783E-22 1.038E-23 0.000E+00
"
- Please also remove the line 124 in FJX_j2j.dat (copied below).
"
123 CH3I PHOTON I PRODUCTS 1.000 /CH3I /
"
--Melissa Sulprizio (talk) 17:44, 27 July 2018 (UTC)