MEGAN v2.1 plus Guenther 2012 biogenic emissions: Difference between revisions
| Line 216: | Line 216: | ||
=== Remove biogenic CO emissions to avoid double counting === | === Remove biogenic CO emissions to avoid double counting === | ||
<span style="color:darkorange">'''''This fix ( | <span style="color:darkorange">'''''This fix (Git ID: [https://github.com/geoschem/geos-chem/commit/8ac8a5cc7e94a28014937cbd9f17f960a814c700 8ac8a5cc]) will be included in [[GEOS-Chem 12#12.6.0|GEOS-Chem 12.6.0]].'''''</span> | ||
'''''Jenny Fisher wrote:''''' | '''''Jenny Fisher wrote:''''' | ||
Revision as of 18:38, 27 August 2019
On this page we describe the MEGAN v2.1 plus Guenther et al 2012 biogenic emissions, which are used in GEOS-Chem v10-01 and newer versions. The MEGAN emissions are fed into GEOS-Chem via the HEMCO emissions component.
Overview
Description
Dylan Millet wrote:
GEOS-Chem emission routines for biogenic VOCs have been updated to MEGAN2.1 as described in Guenther et al., Geosci. Model Dev., 5, 1471–1492, 2012. (Article) The net result is a 11-12% decrease in the global flux for isoprene, and a 30% increase in the global flux for total monoterpenes. Biogenic alkene emissions are now computed using MEGAN instead of by scaling to isoprene, as was done previously. Biogenic emissions of acetaldehyde are now included. Annual emission totals at 2x2.5 resolution for the different compounds are within ~20% of the values in Table 6 of Guenther et al., within the expected uncertainty associated with differing meteorology, years, etc. Note that global emissions can also change by 10% or more when running at 2x2.5 versus 4x5 degree horizontal resolution (for both the updated and the standard emission routines).
One of the ‘under-the-hood’ changes is that there is now a single driver routine for calculating MEGAN emissions across all compounds (GET_MEGAN_EMISSIONS). Previously we had separate driver routines for each individual compound. GET_MEGAN_EMISSIONS is passed the compound name, looks up or computes the appropriate parameters, and returns the corresponding emissions. This should hopefully make it much easier to add new compounds in the future, since one just has to add the appropriate parameter values for that compound. There are parameters included for a number of compounds not presently in the standard chemistry scheme - for use in specialized simulations and/or future inclusion in standard fullchem. Another under-the-hood change is that the PCEEA/PECCA flag is no longer needed since we use this scheme exclusively now.
The new MEGAN implementation computes emissions for certain compounds based on pre-defined emission factor maps provided with the MEGAN source code. For other compounds emissions are computed in GEOS-Chem based on CLM4 plant functional type distributions combined with PFT-specific emission factors. This PFT approach can be used for all MEGAN compounds if desired (by adding in the corresponding emission factors for each PFT), which may be useful for applications using different vegetation maps or dynamically shifting vegetation.
The updated emissions lead to lower CO concentrations in the Northern Hemisphere during summer. A GEOS-Chem v9-01-03 4x5 run for 2006 gave CO mixing ratios averaged for P>800 hPa) that were lower by a few ppb over much of the Northern Hemisphere during ummer months. Maximum decreases of about 10ppb occur over certain source regions. Ox changes are small (< 2ppb for P > 800hPa) using the standard v9-1-3 (non-Caltech) chemistry scheme.
Please contact Dylan Millet with any further questions about these emissions.
--Bob Y. 16:05, 25 February 2015 (EST)
Documentation
- Comparison between previous and updated BVOC emissions for standard tracers (nested NA, year-2011):
- Updated emissions for all BVOCs, including non-standard tracers:
- Monthly CO comparison for previous and updated MEGAN implementation (year-2006; 4x5 resolution):
--Dbm 14:18, 22 January 2013 (EST)
Data files
In GEOS-Chem v10-01 and newer versions, the MEGAN v2.1 plus Guenther (2012) biogenic emissions are read with the HEMCO emissions component. We have created new MEGAN data files (in COARDS-compliant netCDF format) for use with HEMCO. These new data files are contained in the HEMCO data directory tree. For detailed instructions on how to download these data files to your disk server, please see our Downloading the HEMCO data directories wiki post.
--Bob Y. 13:19, 3 March 2015 (EST)
Discussion following 1-month benchmark v10-01h
This emission scheme was validated in GEOS-Chem v10-01h. The following discussion ensued.
Dylan Millet wrote:
- A few comments and questions below.
- 1. Acetone
- Based on the benchmarking I did earlier biogenic acetone emissions should increase 6-7% globally & annually with the MEGAN update. But the emission ratio plot for ACET in the new benchmark shows a >2x increase everywhere. On the emission summary page, the "biogenic and natural' acetone sources look to have increased from 1.6 to 5.2. But if we look on the same page at the "acetone sources" the two simulations are very similar and the sum is consistent with a ~7% increase. In other words the large discrepancy in the emission ratio plot and in the "biogenic and natural sources" table for acetone just looks to be a correction to the way the diagnostics are being done. Is that right? Consistent with that, the concentration ratios are not very different.
- 2. Isoprene
- I'd expect about a 10-12% isoprene decrease globally & annually for isoprene emissions. The new benchmark has a ~30% decrease. I'm not sure why that would be. It could just be the difference between a 1-month run and a full year, or interannual variability, I suppose, though the discrepancy seems large for that. Does the 10-01f benchmark we're comparing to use the (old) standard canopy treatment, or does it use the PCEEA parameterization? That was an option before, but we got rid of it with the MEGAN update (which only uses PCEEA). If we're switching canopy schemes that might explain the difference.
- 3. For propene I expect about a 50% increase globally. The change we see in the benchmark is larger (67%) but not dramatically.
- 4. Since we now have biogenic emissions of ALD2 we should add the appropriate biogenic benchmark diagnostics.
Melissa Sulprizio wrote:
- 1. Acetone
- That is correct. Prior to this version, there was a bug in diag3.F in the conversion to atomsC/cm2/s that affected the biogenic acetone diagnostic (ND46). The error appears to have come in with the implementation of HEMCO. We corrected the unit conversion in this version and the biogenic acetone emissions are now more consistent with pre-HEMCO values.
- 2. Isoprene
- The PECCA/PCEEA model was turned off in the old benchmarks, so that may explain the difference. From the HEMCO.log file for the previous benchmark (v10-01f_UCX), we have:
Use MEGAN biogenic emissions (extension module)
- Use PECCA model: F
- Use the following species:
Isoprene = ISOP 1
Acetone = ACET 26
C3 Alkenes = PRPE 23
Ethene = C2H4 -1
--> Isoprene scale factor is 1.00000000000000
- MEGAN monoterpene option enabled:
CO = CO 31
OC aorosol = OCPI 16
Monoterp. = MONX -1
- 3. Propene
- Could it be that the difference in meteorology fields is causing this difference? It looks like your benchmarks used GEOS-5 met fields for 2006. In v10-01h, we are using GEOS-FP met fields for July 2013.
- 4. Diagnostics
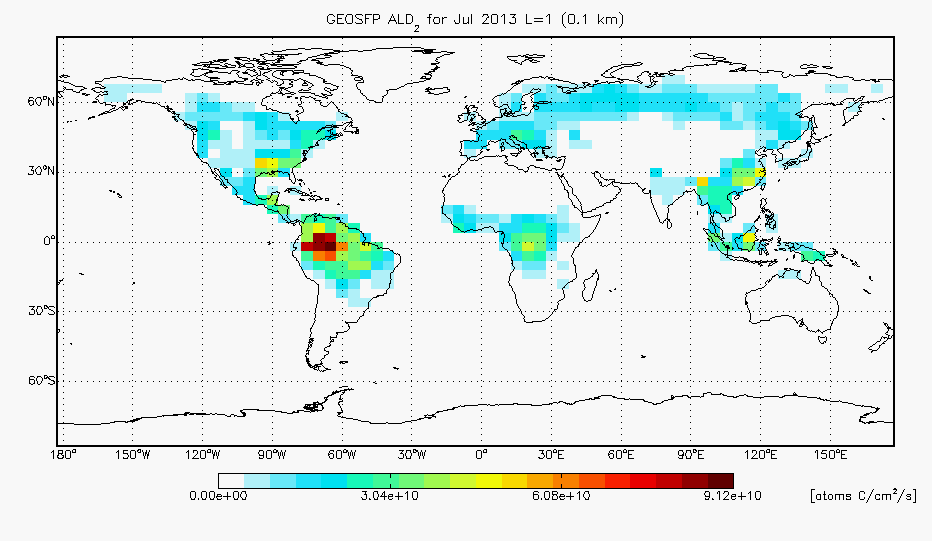
- I have added ALD2 to the benchmark plotting routines. We were unable to make difference plots for this version since v10-01f_UCX didn’t include biogenic emissions of ALD2 so I have that update commented out for now, but we will include differences of ALD2 biogenic emissions in future benchmarks. For your reference, I have included a map of the ALD2 biogenic emissions from v10-01h below:
Dylan Millet wrote:
- Ok, that all seems reasonable. The only question left in my mind is the 30% isoprene decrease compared to 10% that I saw earlier. I've started a run testing the impact of switching canopy schemes to see if that accounts for it.
- Based on my original implementation of the MEGAN update, changing the MEGAN algorithms and EFs gives a 10-11% reduction in global emissions. Changing the canopy scheme gives another 7-8% decrease. So the total expected decrease (18%) is still less than we see with the new benchmark (30%), but close enough that it could be due to 1 month versus 1 year, different met fields, interannual variability, etc. I vote for moving ahead and having a look at the 1-y benchmark.
--Melissa Sulprizio 13:12, 27 March 2015 (EDT)
Update to Yuan processed MODIS LAI data
This update is slated for inclusion in GEOS-Chem 12.3.0.
This section has been moved to our Leaf area indices in GEOS-Chem wiki page.
--Bob Yantosca (talk) 18:59, 8 March 2019 (UTC)
Updated PFT file
This update was included in GEOS-Chem 12.3.0, which was released on 01 Apr 2019.
Jenny Fisher wrote:
Dylan mentioned the updated plant functional type file and higher resolution and suggested this get made part of the standard code. In terms of implementation, the file
HEMCO/MEGAN/v2017-07/CLM4_PFT.geos.025x03125.nc
would be replaced by:
HEMCO/MEGAN/v2018-05/CLM4_PFT.geos.025x03125.v201805.nc
(with appropriate fixes to the HEMCO_Config.rc paths.)
--Melissa Sulprizio (talk) 15:04, 8 August 2018 (UTC)
--Bob Yantosca (talk) 20:42, 11 March 2019 (UTC)
Global 0.25x0.25 MEGAN input and MODIS LAI
This update was included in v11-02f (approved 17 May 2018).
Jenny Fisher wrote:
- The summary version of the issue is:
- Several years back, Mike got a version of the raw MODIS LAI files from BU on a 0.25°x0.25° grid. These had some sort of projection error that led to the weird behaviour I saw over Australia (offsetting along the southern coasts of Australia, w/ some SW ocean grid boxes having LAI > 0 and some SE land boxes having LAI = 0).
- Mike processed those files to a 0.5°x0.5° version for input to GEOS-Chem. This was NOT just regridding the original files, but also making some adjustments following Muller et al. 2008*.
- These were the files in the archive for some unknown period of time.
- Eventually (for HEMCO? Or 0.25x0.3125? or both?) someone must have regridded these 0.5°x0.5° files back to 0.25°x0.25°.
- In the meantime, Eloise received an updated version of the raw MODIS LAI files from BU on a 0.25°x0.25° grid. These did not have the projection error.
- AFAIK, Eloise has not applied the same type of processing Mike did – so what I have now are just the raw data converted to HEMCO format.
- After all of this, my recommendation for moving forward is that we replace the existing files currently used in GEOS-Chem with the versions from Eloise (I now have scripts to prepare these for HEMCO).
Dylan Millet response:
- Using the raw, unscaled LAI files sounds like the way to go for the purposes of other modules. The scaling to the fraction of vegetated area will need to take place inside the megan module. As I recall, since the EFs are already scaled to their fractional coverage, we need to apply scaled LAI (i.e. the LAI for the vegetated area) otherwise we will be underestimating the flux ... in effect double-counting the effect of fractional vegetation coverage.
- I think it would be better to have a single change where both the LAI files and the scaling are taken care of, and also that we should do a quick assessment of how the isoprene flux changes from the prior version, before sending in as a patch for the mainline code.
- The LAI scaling is a simple matter, it just means dividing the LAI by the fraction of the grid square that is covered by vegetation before computing GAMMA_LAI. In other words, MISOLAI needs to be divided by the sum of ARRAY_16(:,:,2:16) prior to GET_GAMMA_LAI in the current version of the code. I guess with a trap to avoid dividing by zero. (Mike, can you confirm that's what happened with the LAI data you produced?) I can certainly work with Christoph to implement (when I get a moment!) but the only thing that requires any thought at all is just the passing of that PFT data into get_megan_emissions as right now it is dealt with in calc_aef.
Christoph Keller response:
- If it's just a matter of normalizing MISOLAI by the PFTs (ARRAY_16) within GET_MEGAN_EMISSIONS then this is a quick fix that I'm happy to provide.
- My only concern would be that the new code should only be used with the updated files. It would be good to make the code stop if the MEGAN module and input files are inconsistent to avoid users mixing up wild combinations of code and data!
Christoph Keller follow-up:
- Attached is a git patch that normalizes MEGAN LAI by PFT. Do you want to do some internal validation / sensitivity analysis before sending it off to the GCST?
- The patch is based of GEOS-Chem v11-01. You should be able to apply it using
git apply Normalize-MEGAN-LAI.patch
- Two comments:
- I did not change the hard-coded path of the LAI files in modis_lai_mod.F90 because it would break my code (I had to set it to a local directory when testing the code).
- I added an optional HEMCO setting to control MEGAN LAI normalization (in HEMCO_Config.rc)
108 MEGAN : on ISOP/ACET/PRPE/C2H4/ALD2
--> Isoprene scaling : 1.0
--> CO2 inhibition : true
--> CO2 conc (ppmv) : 390.0
--> Normalize LAI : false
- You don't have to specify the Normalize LAI setting, it is set to 'true' by default. But this gives you the option to (a) use this version of HEMCO with the old LAI files and (b) test the impact of normalizing the LAI values vs. using the new LAI files without normalization.
--Melissa Sulprizio (talk) 15:32, 20 July 2017 (UTC)
CO2 direct effect on isoprene emissions
This update was validated with 1-month benchmark simulation v11-01d and 1-year benchmark simulation v11-01d-Run1. This version was approved on 12 Dec 2015.
Amos Tai developed code updates to include CO2 inhibition of isoprene emissions in MEGAN.
The reference for this work is:
- Tai, A.P.K., L.J. Mickley, C.L. Heald, S. Wu, Effect of CO2 inhibition on biogenic isoprene emission: Implications for air quality under 2000-to-2050 changes in climate, vegetation, and land use, Geophys. Res. Let., 40, 3479-3483, 2013. [pdf]
--Melissa Sulprizio 15:45, 23 January 2014 (EST)
Note on the use of pre-computed emission factor (EF) maps versus EF maps computed online
As noted earlier on this page, MEGANv2.1 includes precomputed emission factor maps for a subset of VOCs. Emission factors for other VOCs are computed online based on plant functional types from CLM and species-specific emission factors for each of those PFTs. Results from these two approaches can differ significantly. For the current GEOS-Chem implementation the default is to use pre-computed EF maps when available, since for some geographic areas these are based on more detailed plant speciation than can be captured by the broad CLM PFTs. GEOS-Chem species using pre-computed EF maps include ISOP, MBOX, BPIN, CARE, LIMO, OCIM, and SABI. All others use EFs computed online in the model from the PFT data.
In the case of APIN, MYRC, and ACET, pre-computed EF maps were available, but (for the versions available at the time of this implementation), their use led to large emission discrepancies compared to the published Guenther et al. (2012) values. So at the recommendation of Alex Guenther and his group, we use the PFT-EF approach for APIN, MYRC, and ACET rather than the pre-computed maps.
--Dbm (talk) 22:57, 24 November 2018 (UTC)
References
- Barkley, M., Description of MEGAN biogenic VOC emissions in GEOS-Chem, 2010. PDF
- Buermann, W., Wang, Y.J., Dong, J.R., Zhou, L.M., Zeng, X.B., Dickinson, R.E., Potter, C.S., and Myneni, R.B.: Analysis of a multiyear global vegetation leaf area index data set, J. Geophys. Res., 107, 4646, doi:10.1029/2001JD000975, 2002.
- Guenther, A., Baugh, B., Brasseur, G., Greenberg, J., Harley, P., Klinger, L., Serca, D., and Vierling, L.: Isoprene emission estimates and uncertainties for the Central African EXPRESSO study domain, J. Geophys. Res., 104, 30625-30639, 1999.
- Guenther, A., Karl, T., Harley, P., Wiedinmyer, C., Palmer, P.I., and Geron, C.: Estimates of global terrestrial isoprene emissions using MEGAN (Model of Emissions of Gases and Aerosols from Nature), Atmos. Chem. Phys., 6, 3181-3210, 2006.
- Guenther, A., and C. Wiedinmyer, User's guide to the Model of Emissions of Gases and Aerosols from Nature (MEGAN), Version 2.01, 2007.
- Guenther, A. B., Jiang, X., Heald, C. L., Sakulyanontvittaya, T., Duhl, T., Emmons, L. K., and Wang, X.: The Model of Emissions of Gases and Aerosols from Nature version 2.1 (MEGAN2.1): an extended and updated framework for modeling biogenic emissions, Geosci. Model Dev., 5, 1471-1492, doi:10.5194/gmd-5-1471-2012, 2012. Article
- Millet, D.B., Jacob, D.J., Boersma, K.F., Fu, T.M., Kurosu, T.P., Chance, K., Heald, C.L., and Guenther, A.: Spatial distribution of isoprene emissions from North America derived from formaldehyde column measurements by the OMI satellite sensor, J. Geophys. Res., 113, D02307, doi:10.1029/2007JD008950, 2008. PDF
- Mueller, J.-F., et al. Global isoprene emissions estimated using MEGAN, ECMWF analyses and a detailed canopy environment model, Atmos. Chem. Phys., 8, 1329-1341, 2008.
- Myneni, R. B., et al., Large seasonal swings in leaf area of Amazon rainforests, Proceedings of the National Academy of Sciences, 104(12), 4820{4823, doi:10.1073/pnas.0611338104, 2007.
- Palmer, P.I., Abbot, D.S., Fu, T.M., Jacob, D.J., Chance, K., Kurosu, T.P., Guenther, A., Wiedinmyer, C., Stanton, J.C., Pilling, M.J., Pressley, S.N., Lamb, B., and Sumner, A.L.: Quantifying the seasonal and interannual variability of North American isoprene emissions using satellite observations of the formaldehyde column, J. Geophys. Res., 111, D12315, doi:10.1029/2005JD006689, 2006. PDF
- Sakulyanontvittaya, T., T. Duhl, C. Wiedinmyer, D. Helmig, S. Matsunaga, M. Potosnak, J. Milford, and A. Guenther, Monoterpene and Sesquiterpene Emission Estimates for the United States, Environ. Sci. Technol., 42(5), 1623{1629, doi:10.1021/es702274e, 2008.
- Tai, A.P.K., L.J. Mickley, C.L. Heald, S. Wu, Effect of CO2 inhibition on biogenic isoprene emission: Implications for air quality under 2000-to-2050 changes in climate, vegetation, and land use, Geophys. Res. Let., 40, 3479-3483, 2013. [pdf]
--Bob Y. 15:49, 25 February 2015 (EST)
Previous issues that are now resolved
Remove biogenic CO emissions to avoid double counting
This fix (Git ID: 8ac8a5cc) will be included in GEOS-Chem 12.6.0.
Jenny Fisher wrote:
- [CO in MEGAN was included] to account for CO chemical production from monoterpenes that were not previously included in the chemical mechanism. Now that we include them & their chemistry explicitly (right?) I would think that would mean we wouldn’t need to include an extra yield of CO (or if we did it would be smaller because it would only represent species we still don’t include).
Melissa Sulprizio wrote:
- In v11-02c, we added updated isoprene and monoterpene chemistry based on Travis et al. (2016) and Fisher et al. (2016). In the current full-chemistry mechanism (see Standard.eqn), we do include chemistry of MTPA, MTPO, and LIMO. For example:
MTPA + O3 = 0.850OH + 0.100HO2 +
0.620KO2 + 0.140CO + 0.020H2O2 +
0.650RCHO + 0.530MEK : GCARR(5.00E-16, 0.0E+00, -530.0); {2017/07/14; Atkinson2003; KRT,JAF,CCM,EAM,KHB,RHS}
MTPO + O3 = 0.850OH + 0.100HO2 +
0.620KO2 + 0.140CO + 0.020H2O2 +
0.650RCHO + 0.530MEK : GCARR(5.00E-16, 0.0E+00, -530.0); {2017/07/14; Atkinson2003; KRT,JAF,CCM,EAM,KHB,RHS}
MTPA + NO3 = 0.100OLNN + 0.900OLND : GCARR(8.33E-13, 0.0E+00, 490.0); {2017/07/14; Fisher2016; KRT,JAF,CCM,EAM,KHB,RHS}
MTPO + NO3 = 0.100OLNN + 0.900OLND : GCARR(8.33E-13, 0.0E+00, 490.0); {2017/07/14; Fisher2016; KRT,JAF,CCM,EAM,KHB,RHS}
LIMO + OH = LIMO2 : GCARR(4.20E-11, 0.0E+00, 401.0); {2017/07/14; Gill2002; KRT,JAF,CCM,EAM,KHB,RHS}
LIMO + O3 = 0.850OH + 0.100HO2 +
0.160ETO2 + 0.420KO2 + 0.020H2O2 +
0.140CO + 0.460PRPE + 0.040CH2O +
0.790MACR + 0.010HCOOH + 0.070RCOOH : GCARR(2.95E-15, 0.0E+00, -783.0); {2017/07/14; Atkinson2003; KRT,JAF,CCM,EAM,KHB,RHS}
LIMO + NO3 = 0.500OLNN + 0.500OLND : GCARR(1.22E-11, 0.0E+00, 0.0); {2017/07/14; Fry2014,Atkinson2003; KRT,JAF,CCM,EAM,KHB,RHS}
- In the MEGAN HEMCO extension, we also have the following:
! CO from MONOTERPENE oxidation (bnd, bmy, 1/2/01)
!
! Assume the production of CO from monoterpenes is instantaneous
! even though the lifetime of intermediate species may be on the
! order of hours or days. This assumption will likely cause CO from
! monoterpene oxidation to be too high in the box in which the
! monoterpene is emitted.
!
! The CO yield here is taken from:
!
! Hatakeyama et al. JGR, Vol. 96, p. 947-958 (1991)
! "The ultimate yield of CO from the tropospheric oxidation of
! terpenes (including both O3 and OH reactions) was estimated
! to be 20% on the carbon number basis." They studied
! ALPHA- & BETA-pinene.
!
! Vinckier et al. Fresenius Env. Bull., Vol. 7, p.361-368 (1998)
! "R(CO)=1.8+/-0.3" : 1.8/10 is about 20%.
!
! EMIS_MONO is now kgC, conversion factor then becomes:
! 28g CO / 60g C = 46.666%
! (ckeller, 4/15/14).
!--------------------------------------------------------------------
! NOTE: Don't save into EMISRR for the tagged CO simulation.
! Also for tagged CO we don't use monoterpenes ??????
! (jaf, mak, bmy, 2/14/08)
IF ( (Inst%IDTCO>0) .AND. (Inst%ExtNrMono>0) ) THEN
!FLUX = EMMO * 0.2d0
FLUXCO(I,J) = EMIS_MONO * 0.4666_hp
ENDIF
- Therefore, biogenic CO emissions are calculated whenever CO is passed to that extension, as is the case in the full-chemistry simulations. In the HEMCO_Config.rc for full-chemistry simulations, we have:
109 MEGAN_Mono : on CO/MTPA/MTPO/LIMO/SESQ
- That does not appear to be the case in tagged CO simulations. From HEMCO_Config.rc for tagCO:
109 MEGAN_Mono : on MTPA/LIMO/MTPO
- When we switched to the offline biogenic VOC emissions in GEOS-Chem 12.4.0, biogenic CO emissions were saved out at high-resolution from the MEGAN extension and we now read those in directly via HEMCO, along with the other offline biogenic VOC emissions. However, if it is in fact a bug that CO emissions in the MEGAN extension are turned on for full-chemistry simulations because it is double-counting that source, then a simple fix would be to remove the offline biogenic CO emissions (and online MEGAN CO emissions).
--Melissa Sulprizio (talk) 18:36, 27 August 2019 (UTC)
Grid offset bug fix in high-resolution MEGAN files at southern mid-latitudes
This fix was implemented in GEOS-Chem 12.0.0.
Jenny Fisher wrote:
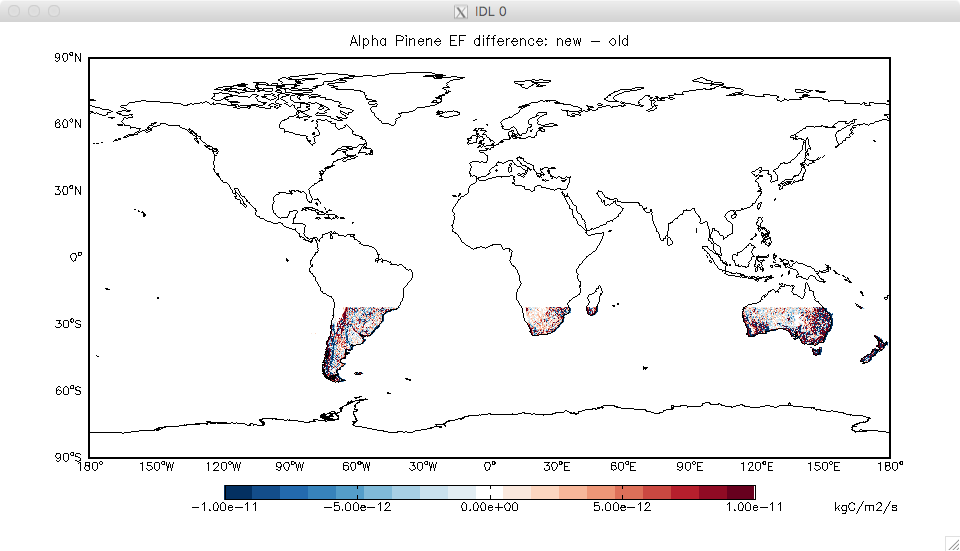
- I’ve discovered a bug in some of the high-resolution files used for MEGAN (previously provided in the MEGAN/v2017-07/). For one species (alpha pinene) there was a strange offset of the grid that affected the southern mid-latitudes. With Dylan’s help, we traced this to the original very high resolution MEGAN input files provided by Alex Guenther’s group. These were updated in Nov 2016, removing the offset.
- I’ve recreated the HEMCO-ready version of these files, and they are available for download. This is definitely a bug fix, so should get implemented during the v11-02 provisional release period (really just a file update so it’s easy). Note that the update will ONLY affect anyone looking at monoterpenes (specifically MTPA) in the southern mid-latitudes – see attached difference plot.
The new files are stored in ExtData/HEMCO/MEGAN/v2018-05/ with an accompanying README.
--Lizzie Lundgren (talk) 14:55, 31 May 2018 (UTC)
Bug fix for monoterpenes in ND46 diagnostic
This fix was validated with the 1-month benchmark simulation v11-01c and approved on 14 Sept 2015.
Jared Brewer wrote:
- I'm now working with the public release of GEOS-Chem v10-01, and I'm reaching out to you regarding a possible bug that I've identified in the monoterpene outputs in the BIOGSRCE diagnostic in diag3.F - specifically the FACTOR code in line 4028. At this line, the program claims it is converting from KgC/m2/s to AtomsC/cm2/s, but includes the factor of 10 for conversion from Kg monoterpene to atoms C (IE, 10 carbons/monoterpene). This bug would appear to be common in all the different monoterpene emissions as well (A-Pinene, Ocimene, etc.).
To resolve this issue, we have removed the number of carbons per molecume (10 for monoterpenes, 15 for sesquiterpenes, etc.( from the conversion factors in diag3.F.
--Melissa Sulprizio (talk) 18:36, 9 September 2015 (UTC)
Minor bug fix in MEGAN_Mono extension
This update was added as a last-minute fix to GEOS-Chem v10-01. It was included in the GEOS-Chem v10-01 public release (17 Jun 2015).
Christoph Keller wrote:
- I came across a small bug in MEGAN: I tried to run the "standard" MEGAN only, i.e. without the monoterpenes, but this would give me zeros for all emissions except isoprene because the CLM4 arrays were only being read if MEGAN extension 109 (MEGAN_mono) was enabled. So in the configuration file, we need to link the CLM4 arrays to extension 108, not 109:
108 CLM4_PFT_BARE $ROOT/MEGAN/v2015-02/CLM4_PFT.geos.1x1.nc PFT_BARE 2000/1/1/0 C xy 1 * - 1 1
- etc. Furthermore, I made two small updates in HEMCO/Extensions/hcox_megan_mod.F for better error trapping and to avoid out-of-bounds errors:
- 1. In the run driver (HCOX_Megan_Run), evaluate the error return code of CALC_AEF and CALC_NORM_FAC:
IF ( FIRST ) THEN
! Generate annual emission factors for MEGAN inventory
CALL CALC_AEF( am_I_Root, HcoState, ExtState, RC )
IF ( RC /= HCO_SUCCESS ) RETURN
! Calculate normalization factor (dbm, 11/2012)
CALL CALC_NORM_FAC( am_I_Root, RC )
IF ( RC /= HCO_SUCCESS ) RETURN
ENDIF
- 2. In routine CALC_AEF, check if arrays are really used:
! Convert AEF arrays to [kgC/m2/s]
! Multiply arrays by FACTOR and ratio [g C/g compound]
! NOTE: AEFs for ISOP, MBOX, BPIN, CARE, LIMO, OCIM, SABI
! are read from file in [kgC/m2/s], so no need to convert here
IF ( ExtNrMono > 0 ) THEN
AEF_APIN(I,J) = AEF_APIN(I,J)
& * FACTOR * 120.0_hp / 136.234_hp
AEF_MYRC(I,J) = AEF_MYRC(I,J)
& * FACTOR * 120.0_hp / 136.234_hp
AEF_OMON(I,J) = AEF_OMON(I,J)
& * FACTOR * 120.0_hp / 136.234_hp
ENDIF
- Note that these fixes do not affect the benchmarks since the benchmarks run with MEGAN_Mono turned on.
--Bob Y. 16:54, 28 April 2015 (EDT)
Restore missing BIOGENIC_CO diagnostics
This update was added as a last-minute fix to GEOS-Chem v10-01. It was included in the GEOS-Chem v10-01 public release (17 Jun 2015).
Dylan Millet wrote:
CO SOURCES
Tracer v10-01e-geosfp-Run1 v10-01h-geosfp-Run0 v10-01i-geosfp-Run0
============================================================================
COan+bf 527.122265 598.575584 608.052597 Tg
CObb 283.579062 296.993630 293.039311 Tg
COmono 34.826250 41.440323 0.000000 Tg
- Why did the CO source from terpenes change [in GEOS-Chem v10-01]? It looks like it went to zero? Is that just a change in the diagnostic?
Bob Yantosca replied:
- The zero CO from monoterpenes is a diagnostic issue. I looked through the logs from a recent simulation and I saw:
HEMCO WARNING: Diagnostics counter is zero - return empty array: BIOGENIC_CO --> LOCATION: DiagnCont_PrepareOutput (hco_diagn_mod.F90)
- Melissa and I just traced this to an omission in the HEMCO MEGAN extension (module HEMCO/Extensions/hcox_megan_mod.F). HEMCO was computing the CO emissions from monoterpenes properly. But these emissions were not being added to the proper diagnostic container (called BIOGENIC_CO) in HEMCO’s data structure. Therefore, when we went to print out these emissions, they showed up as all zeroes.
- It appears that the subroutine call to add the CO emissions from monoterpenes to the diagnostics had been somehow removed from the GEOS-Chem v10-01i code. We think that this may have been “clobbered” by a recent Git merge. In any case, adding the lines in GREEN correct this issue:
! ----------------------------------------------------------------
! CO
IF ( ( ExtNrMono > 0 ) .AND. ( IDTCO > 0 ) ) THEN
! Add flux to emission array
CALL HCO_EmisAdd( am_I_Root, HcoState, FLUXCO, IDTCO,
& RC, ExtNr=ExtNr )
IF ( RC /= HCO_SUCCESS ) THEN
CALL HCO_ERROR( 'HCO_EmisAdd error: FLUXCO', RC )
RETURN
ENDIF
! Also archive the BIOGENIC_CO diagnostic (bmy, 4/29/15)
Arr2D => FLUXCO
DiagnName = 'BIOGENIC_CO'
CALL Diagn_Update( am_I_Root, ExtNr=ExtNrMono,
& cName=TRIM(DiagnName), Array2D=Arr2D, RC=RC)
IF ( RC /= HCO_SUCCESS ) RETURN
Arr2D => NULL()
ENDIF
- With the fix installed, HEMCO now successfully adds the CO emitted from monoterpenes to the BIOGENIC_CO diagnostic instead of printing out all zeroes.
--Bob Y. 12:01, 29 April 2015 (EDT)