TOMAS aerosol microphysics: Difference between revisions
m (I corrected my email and added my institute to the list of users.) |
|||
| (261 intermediate revisions by 8 users not shown) | |||
| Line 1: | Line 1: | ||
This page describes the TOMAS aerosol microphysics option in GEOS-Chem. TOMAS is one of two aerosol microphysics packages being incorporated into GEOS-Chem, the other being [[APM aerosol microphysics|APM]]. | |||
== Overview == | == Overview == | ||
The TwO-Moment Aerosol Sectional (TOMAS) microphysics package was developed for implementation into GEOS-Chem at Carnegie-Mellon University. Using a moving sectional and moment-based approach, TOMAS tracks two independent moments (number and mass) of the aerosol size distribution for | The TwO-Moment Aerosol Sectional (TOMAS) microphysics package was developed for implementation into GEOS-Chem at Carnegie-Mellon University. Using a moving sectional and moment-based approach, TOMAS tracks two independent moments (number and mass) of the aerosol size distribution for a number of discrete size bins. It also contains codes to simulate nucleation, condensation, and coagulation processes. The aerosol species that are considered with high size resolution are sulfate, sea-salt, OC, EC, and dust. An advantage of TOMAS is the full size resolution for all chemical species and the conservation of aerosol number, the latter of which allows one to construct aerosol and CCN number budgets that will balance. | ||
=== Authors and collaborators === | === Authors and collaborators === | ||
* [mailto:petera@andrew.cmu.edu Peter Adams] ''(Carnegie-Mellon U.)'' -- Principal Investigator | * [mailto:petera@andrew.cmu.edu Peter Adams] ''(Carnegie-Mellon U.)'' -- Principal Investigator | ||
* [mailto: | * [mailto:win.t@chula.ac.th Win Trivitayanurak] ''(Chulalongkorn University, Thailand)'' | ||
* [mailto: | * [mailto:danielmw@princeton.edu Dan Westervelt] ''(Princeton University, formerly Carnegie-Mellon U.)'' | ||
* [mailto:jeffrey.pierce@ | * [mailto:jeffrey.pierce@colostate.edu Jeffrey Pierce] ''(CSU/Dalhousie U.)'' | ||
* [mailto:jkodros@atmos.colostate.edu Jack Kodros] ''(CSU)'' | |||
* [mailto:sal.farina@gmail.com Salvatore Farina] ''(Colorado State U.)'' | |||
*Marguerite Marks (CMU) | |||
--[[User:Dan Westervelt|Dan W.]] 11:53, 27 January 2010 (EST) | --[[User:Dan Westervelt|Dan W.]] 11:53, 27 January 2010 (EST) | ||
| Line 22: | Line 26: | ||
|-valign="top" | |-valign="top" | ||
|[http://www.ce.cmu.edu/%7Eadams/ Carnegie-Mellon University] | |[http://www.ce.cmu.edu/%7Eadams/ Carnegie-Mellon University] | ||
|[http://www.ce.cmu.edu/%7Eadams/people.htm#peter Peter Adams]<br>[ | |[http://www.ce.cmu.edu/%7Eadams/people.htm#peter Peter Adams]<br>[http://www.ce.cmu.edu/~dwesterv/Site/Home.html Dan Westervelt] | ||
| ... | | [http://www.atmos-chem-phys-discuss.net/13/8333/2013/acpd-13-8333-2013.html New particle formation evaluation in GC-TOMAS] <br> Sensitivity of CCN to nucleation rates <br> Development of number tagging and source apportionment model for GC-TOMAS | ||
|-valign="top" | |||
|[http://www.atmos.colostate.edu/faculty/pierce.php Colorado State] | |||
|[http://pierce.atmos.colostate.edu/ Jeffrey Pierce]<br>Sal Farina<br>[http://www.engr.colostate.edu/~jkodros/index.html Jack Kodros] | |||
|Sensitivity of CCN to condensational growth rates <br> TOMAS parallelization <br> Aerosol radiative effects | |||
|-valign="top" | |||
|[http://https://env.eng.chula.ac.th/ Chulalongkorn University] | |||
|Win Trivitayanurak | |||
|Source apportionment for Southeast Asia <br>Improving size fraction information for emissions <br>Development of WRF-GC-TOMAS | |||
|-valign="top" | |-valign="top" | ||
| | |Add yours here | ||
| | | | ||
| | | | ||
|} | |} | ||
--[[User:Bmy|Bob Y.]] 16:35, 12 May 2014 (EDT) | |||
--[[User:wint|Win T.]] 4:24, 2 Nov 2024 (GMT+7) | |||
TOMAS | == TOMAS-specific setup == | ||
TOMAS has its own run directories (run.Tomas) that can be downloaded from the Harvard FTP. The <tt>input.geos</tt> file will look slightly different from standard GEOS-Chem, and between versions. | |||
Pre- v9.02: | |||
To turn on TOMAS, see the "Microphysics menu" in <tt>input.geos</tt> and make sure TOMAS is set to '''T'''. | |||
--[[User: | v9.02 and later: | ||
TOMAS is enabled or disabled at compile time - the TOMAS flag in input.geos has been removed. | |||
TOMAS is a simulation type 3 and utilizes 171-423 tracers. Each aerosol species requires 30 tracers for the 30 bin size resolution, 12 for the 12 bin, etc. Here is the (abbreviated) default setup in input.geos for TOMAS-30 in v9.02 and later (see run.Tomas directory): | |||
Tracer # Description | |||
1- 62 Std Geos Chem | |||
63 H2SO4 | |||
64- 93 Number | |||
94-123 Sulfate | |||
124-153 Sea-salt | |||
154-183 Hydrophilic EC | |||
184-213 Hydrophobic EC | |||
214-243 Hydrophilic OC | |||
244-273 Hydrophobic OC | |||
274-303 Mineral dust | |||
304-333 Aerosol water | |||
TOMAS-40 requires 423 tracers (~360 TOMAS tracers for each of the 40-bin species, and ~62 standard GEOS-Chem tracers) | |||
--[[User:Salvatore Farina|Salvatore Farina]] 18:48, 8 July 2013 (EDT) | |||
== Implementation notes == | == Implementation notes == | ||
TOMAS | #The original implementation and validation of TOMAS had been done for version [[GEOS-Chem v8-03-01]], which was released on 24 Feb 2010. | ||
#TOMAS was completely re-integrated into [[GEOS-Chem v9-02]], which was released on 03 Mar 2014. | |||
#TOMAS was re-integrated into [[GEOS-Chem v10-01]] to become compatible with HEMCO. | |||
#Updates to TOMAS to use the SOAP/SOAS tracers were made for [[GEOS-Chem v11-02]] (not yet released). | |||
=== | === Update April 2013 === | ||
<span style="color:green">'''''This update was tested in the 1-month benchmark simulation [[GEOS-Chem_v9-02_benchmark_history#v9-02k|v9-02k]] and approved on 07 Jun 2013.'''''</span> | |||
Sal Farina has been working with the GEOS-Chem Support Team to inline the TOMAS aerosol microphysics code into the <tt>GeosCore</tt> directory. All TOMAS-specific sections of code are now segregated from the rest of GEOS-Chem with C-preprocessor statements such as: | |||
#if defined( TOMAS ) | |||
# if defined( TOMAS40 ) | |||
... Code for 40 bin TOMAS simulation (optional) goes here ... | |||
# elif defined( TOMAS12 ) | |||
... Code for 12 bin TOMAS simulation (optional) goes here ... | |||
# elif defined( TOMAS15 ) | |||
... Code for 15 bin TOMAS simulation (optional) goes here ... | |||
# else | |||
... Code for 30 bin TOMAS simulation (default) goes here ... | |||
# endif | |||
#endif | |||
TOMAS is now invoked by compiling GEOS-Chem with one of the following options: | |||
{| border=1 cellspacing=0 cellpadding=5 | |||
|- bgcolor="#cccccc" | |||
!width="250px"|Command | |||
!width="450px"|Result | |||
|-valign="top" | |||
|<tt>make -j4 TOMAS=yes ...</tt> | |||
|Compiles GEOS-Chem for the 30 bin (default) TOMAS simulation | |||
|-valign="top" | |||
|<tt>make -j4 TOMAS12=yes ...</tt> | |||
|Compiles GEOS-Chem for the 12 bin (optional) TOMAS simulation | |||
|-valign="top" | |||
|<tt>make -j4 TOMAS15=yes ...</tt> | |||
|Compiles GEOS-Chem for the 15 bin (optional) TOMAS simulation | |||
|-valign="top" | |||
|<tt>make -j4 TOMAS40=yes ...</tt> | |||
|Compiles GEOS-Chem for the 40 bin (optional) TOMAS simulation | |||
|} | |||
The <tt>-j4</tt> in the above examples tell the GNU Make utility to compile 4 files at a time. This reduces the overall compilation time. | |||
All files in the old <tt>GeosTomas/</tt> directory have now been deleted, as these have been rendered obsolete. | |||
These updates are included in [[GEOS-Chem v9-02]]. These modifications will not affect the existing GEOS-Chem simulations, as all TOMAS code is not compiled into the executable unless you compile with one of the TOMAS options (described in the above table) at compile time. | |||
--[[User:Salvatore Farina|Salvatore Farina]] 13:49, 4 June 2013 (EDT)<br> | |||
--[[User:Bmy|Bob Y.]] 16:39, 12 May 2014 (EDT) | |||
== Computational Information == | |||
GC-TOMAS v9-02 (30 sections) on 8 processors: | |||
*One year simulation = 7-8 days wall clock time | |||
More speedups are available using lower aerosol size resolution | |||
--[[User:Dan Westervelt|Dan W.]] 11:00, 07 May 2013 (EST) | |||
GC-TOMAS v9-02 on 16 processors (glooscap) | |||
{| border=1 cellspacing=0 cellpadding=5 | |||
|- bgcolor="#cccccc" | |||
!width="150px"|Simulation | |||
!width="200px"|Wall time / simulation year | |||
|- | |||
|TOMAS12 (optional) | |||
|2.8 days | |||
|- | |||
|TOMAS15 (optional) | |||
|3.3 days | |||
|- | |||
|TOMAS30 (default) | |||
|6.1 days | |||
|- | |||
|TOMAS40 (optional) | |||
|7.8 days | |||
|} | |||
--[[User:Salvatore Farina|Salvatore Farina]] 15:51, 3 March 2014 (EST) | |||
== Microphysics Code== | |||
The aerosol microphysics code is largely contained within the file <tt>tomas_mod.f</tt>. Tomas_mod and its subroutines are modular -- they use all their own internal variables. For details, see tomas_mod.f and comments. | |||
=== Nucleation === | |||
The choice of nucleation theory is selected in the header section of <tt>tomas_mod.f</tt>. The choices are currently binary homogeneous nucleation as in Vehkamaki, 2001 or ternary homogenous nucleation as in Napari et al., 2002. The ternary nucleation rate is typically scaled by a globally uniform tuning factor of 10^-4 or 10^-5. Binary nucleation (Vehkamaki et al. 2002), ion-mediated nucleation (Yu, 2008) and activation nucleation (Kulmala, 2006) are options as well. | |||
In TOMAS-12 and TOMAS-30, nucleated particles follow the Kerminen approximation to grow to the smallest size bin. This has a tendency to overpredict the number of particles in the smallest bins of those models. See Y. H. Lee, J. R. Pierce, and P. J. Adams 2013 [http://www.geosci-model-dev-discuss.net/6/893/2013/gmdd-6-893-2013.html here] for more details on the consequences of this. | |||
=== Condensation === | |||
=== Coagulation === | |||
--[[User:Dan Westervelt|Dan W.]] 14:08, 9 May 2011 (EST) | |||
== Validation == | |||
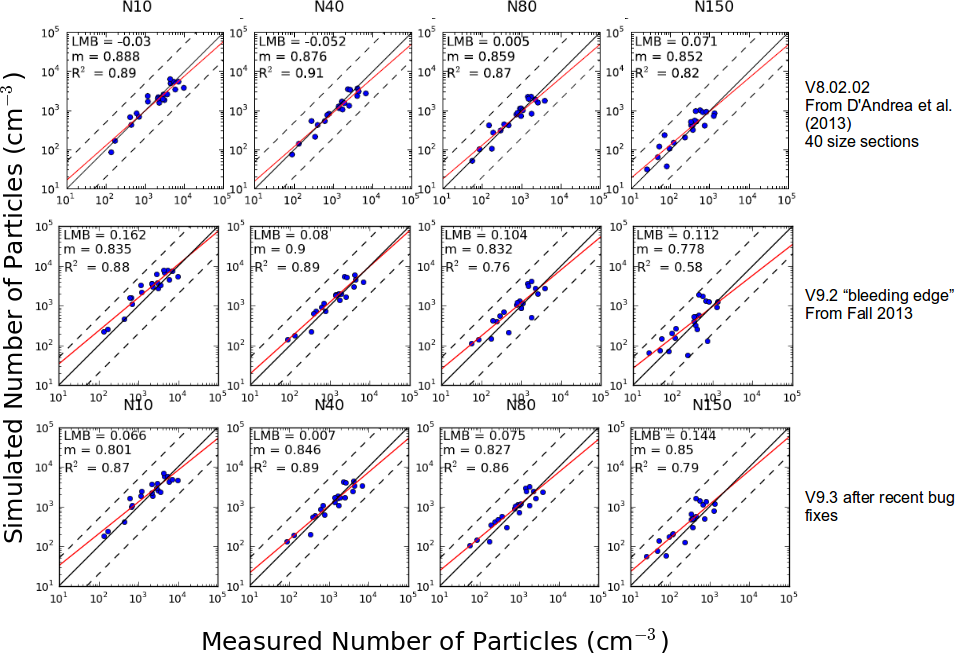
The following figure documents the performance of GEOS-Chem-TOMAS for predicting aerosol number (N10 = number of particles larger than 10 nm etc.) against measurements at 20 global sites. Details of observations are in | |||
* D'Andrea, S. D., Hakkinen, S. A. K., Westervelt, D. M., Kuang, C., Levin, E. J. T., Kanawade, V. P., Leaitch, W. R., Spracklen, D. V., Riipinen, I., and Pierce, J. R.: ''Understanding global secondary organic aerosol amount and size-resolved condensational behavior'', <u>Atmos. Chem. Phys.</u>, '''13''', 11519-11534, doi:10.5194/acp-13-11519-11534, 2013. | |||
[[Image:GEOS-Chem-TOMAS performance update 20140304.png]] | |||
An updated version of this figure is included as Figure 1 in Kodros and Pierce, (2017). Please note that the figure in this paper uses GEOS-Chem v10-01, which included substantial updates to emission inventories (as part of the HEMCO update). Thus, the differences in the comparisons between GEOS-Chem v8-02, v9-03, and v10-01 shown here are not necessarily due to the TOMAS code alone. | |||
- | *Kodros, J. K. and Pierce, J. R.: Important global and regional differences in aerosol cloud-albedo effect estimates between simulations with and without prognostic aerosol microphysics, J. Geophys. Res. Atmos., doi:10.1002/2016JD025886, 2017 | ||
--[[User:Jeff Pierce|Jeff Pierce]] 13:21, 4 March 2014 (MST) <br> | |||
--[[User:Jkodros|Jack Kodros]] 11:30, 11 June 2018 (MST) | |||
== Other features of TOMAS == | |||
Other varieties of TOMAS are suited for specific science questions, for example with nucleation studies where explicit aerosol dynamics are needed for nanometer-sized particles. | |||
=== Set-up Guide === | |||
This [[TOMAS setup guide]] was written for users on ACE-NET's Glooscap cluster, but may be more generally applicable. | |||
--[[User: | --[[User:Salvatore Farina|Salvatore Farina]] 11:55, 26 July 2013 (EDT) | ||
== | === Size Resolution === | ||
The different TOMAS simulations (12, 15, 30, 40 size bins) have the following characteristics: | |||
The | |||
-- | {| border=1 cellspacing=0 cellpadding=5 | ||
|- bgcolor="#cccccc" | |||
!width="100px"|Simulation | |||
!width="700px"|Size resolution | |||
|-valign="top" | |||
|TOMAS12 | |||
|All 7 chemical species have size resolution ranging from 10 nm to 1 µm spanned by 10 logarithmically spaced (mass quadrupling) bins and two supermicron bins. Coarser resolution than TOMAS30 - Improved computation time. | |||
|-valign="top" | |||
|TOMAS15 | |||
|Same as TOMAS12 with 3 additional (mass quadrupling) sub-10nm bins with a lower limit ~2nm. Analogous to TOMAS40 with improved computation time. | |||
|-valign="top" | |||
|TOMAS30 | |||
|All 7 chemical species have size resolution ranging from 10 nm to 10 µm, spanned by 30 logarithmically spaced (mass doubling) bins. | |||
|-valign="top" | |||
|TOMAS40 | |||
|Same as TOMAS30 with 10 additional (mass doubling) sub-10nm bins with a lower limit ~1nm. | |||
|} | |||
--[[User:Salvatore Farina|Salvatore Farina]] 12:51, 4 June 2013 (EDT) | |||
=== Nesting and grid size === | |||
As of v10.01, TOMAS has been implemented and tested on the 4x5, 2x2.5, 0.5x0.667 (North America and Asia), and 0.25x0.3125 (Asian) domains. To the best of our knowledge, these grids should continue working in upcoming releases of GEOS-Chem (including the grid-independent GEOS-Chem). | |||
=== Sample code === | |||
To assist new users, sample processing code (in Python) is available for GEOS-Chem-TOMAS output. Specifically, this code reads in GEOS-Chem-TOMAS output and demonstrates some common calculations (such as calculating size distributions, bin diameters, CCN, etc.). The primary focus of this code is to provide a few examples rather than a complete, efficient package. Further, this code was written based on GEOS-Chem-TOMAS v10.01, which outputs monthly tracers in bpch format. We also provide sample IDL scripts to convert bpch files to netCDF using GAMAP routines. Future versions of GEOS-Chem will output netCDF directly, and so these IDL routines will become obsolete. | |||
Git repository for sample TOMAS code: https://bitbucket.org/teampierce/sample_processing/src/master/ | |||
--[[User:Jkodros|Jack Kodros]] 1:12, 11 June 2018 (MST) | |||
--- | === AOD, CCN post-processing code === | ||
Codes available for calculating aerosol optical depth using TOMAS predicted aerosol composition and size and Mie Theory. Also CCN concentrations calculated from TOMAS size-resolved composition and Kohler theory. Developed by Yunha Lee and Jeffrey Pierce, adapted for GEOS-Chem output by Jeffrey Pierce. | |||
[[ | --[[User:Dan Westervelt|Dan W.]] 2:00, 9 May 2011 (EST) | ||
=== Biomass burning subgrid coagulation switch === | |||
<span style="color:green">'''''This update was included in [[GEOS-Chem_12#12.2.1|GEOS-Chem 12.2.1]], which was released on 28 Feb 2019.'''''</span> | |||
Emily Ramnarine created code that allows the "emitted" size distribution in the model be a function of a number of properties that include the mean emissions rate per fire in the grid box. In order to do this, Emily needed to (1) make changes to carbon_mod.F, (2) add a line or few to HEMCO_config.rc, and (3) modify the FINN input files to include the number of fires. This only affects TOMAS simulations. Emily wrote: | |||
:This parameterization, based on Sakamoto et al (2016), estimates the amount of near-source, sub-grid scale coagulation happening in a biomass burning plume. Can be turned on or off. When on, the default assumption is that each smoke plume is completely seperate from the others (i.e. there is no overlap of the plumes). There is also an option for all smoke plumes in a grid box to overlap completely. When being used, this parameterization changes the median diameter and modal width of biomass burning emissions to account for coagulation. | |||
- | '''Reference''' | ||
Ramnarine, E., Kodros, J. K., Hodshire, A. L., Lonsdale, C. R., Alvarado, M. J., and Pierce, J. R., ''Effects of Near-Source Coagulation of Biomass Burning Aerosols on Global Predictions of Aerosol Size Distributions and Implications for Aerosol Radiative Effects'', <u>Atmos. Chem. Phys. Discuss.</u>, https://doi.org/10.5194/acp-2018-1084, in review, 2018. | |||
--[[User:Melissa Payer|Melissa Sulprizio]] ([[User talk:Melissa Payer|talk]]) 15:07, 22 February 2019 (UTC)<br>--[[User:Bmy|Bob Yantosca]] ([[User talk:Bmy|talk]]) 18:54, 28 February 2019 (UTC) | |||
=== | == References == | ||
In this section we provide references relevant to TOMAS aerosl microphysics simulations. | |||
--[[User: | === Studies using TOMAS simulations === | ||
#'''Nucleation in GEOS-Chem:''' Westervelt, D. M., Pierce, J. R., Riipinen, I., Trivitayanurak, W., Hamed, A., Kulmala, M., Laaksonen, A., Decesari, S., and Adams, P. J.: ''Formation and growth of nucleated particles into cloud condensation nuclei: model-measurement comparison'', <u>Atmos. Chem. Phys. Discuss.</u>, '''13''', 8333-8386, doi:10.5194/acpd-13-8333-2013, 2013. [http://www.atmos-chem-phys-discuss.net/13/8333/2013/acpd-13-8333-2013.html LINK] | |||
#'''TOMAS implementation in GEOS-Chem:''' Trivitayanurak, W., Adams, P. J., Spracklen, D. V. and Carslaw, K. S.: ''Tropospheric aerosol microphysics simulation with assimilated meteorology: model description and intermodel comparison'', <u>Atmos. Chem. Phys.</u>, '''8'''(12), 3149-3168, 2008. | |||
#'''TOMAS initial paper, sulfate only:''' Adams, P. J. and Seinfeld, J. H.: ''redicting global aerosol size distributions in general circulation models'', <u>J. Geophys. Res.-Atmos.</u>, '''107'''(D19), -, doi:Artn 4370 Doi 10.1029/2001jd001010, 2002. | |||
#'''TOMAS with sea-salt:''' Pierce, J.R., and Adams P.J., ''Global evaluation of CCN formation by direct emission of sea salt and growth of ultrafine sea salt'', <u>J. Geophys. Res.-Atmos.</u>, '''111''' (D6), doi:10.1029/2005JD006186, 2006. | |||
#'''TOMAS with carbonaceous aerosol:''' Pierce, J. R., Chen, K. and Adams, P. J.: ''Contribution of primary carbonaceous aerosol to cloud condensation nuclei: processes and uncertainties evaluated with a global aerosol microphysics model'', <u>Atmos. Chem. Phys.</u>, '''7'''(20), 5447-5466, doi:10.5194/acp-7-5447-2007, 2007.; Trivitayanurak, W. and Adams, P. J.: ''Does the POA–SOA split matter for global CCN formation?'', <u>Atmos. Chem. Phys.</u>, '''14''', 995–1010, doi:10.5194/acp-14-995-2014, 2014. | |||
#'''TOMAS with dust:''' Lee, Y.H., K. Chen, and P.J. Adams, 2009: ''Development of a global model of mineral dust aerosol microphysics''. <u>Atmos. Chem. Phys.</u>, '''8''', 2441-2558, doi:10.5194/acp-9-2441-2009. | |||
#'''TOMAS with SOA:''' D'Andrea, S. D., Hakkinen, S. A. K., Westervelt, D. M., Kuang, C., Levin, E. J. T., Kanawade, V. P., Leaitch, W. R., Spracklen, D. V., Riipinen, I., and Pierce, J. R.: ''Understanding global secondary organic aerosol amount and size-resolved condensational behavior'', <u>Atmos. Chem. Phys.</u>, '''13''', 11519-11534, doi:10.5194/acp-13-11519-11534, 2013. | |||
#'''TOMAS with offline DRE/AIE:''' Kodros, J. K., Cucinotta, R., Ridley, D. A., Wiedinmyer, C. and Pierce, J. R.: ''The aerosol radiative effects of uncontrolled combustion of domestic waste'', <u>Atmos. Chem. Phys.</u>, 16(11), 6771-6784, doi:10.5194/acp-16- 6771-2016, 2016 | |||
#'''TOMAS compared to GEOS-Chem standard:''' Kodros, J. K. and Pierce, J. R.: ''Important global and regional differences in aerosol cloud-albedo effect estimates between simulations with and without prognostic aerosol microphysics'', <u>J. Geophys. Res. Atmos.</u>, doi:10.1002/2016JD025886, 2017. | |||
--[[User:Bmy|Bob Y.]] 09:53, 2 June 2014 (EDT), [[User:Wint|Win T.]] 17:18, 28 June 2021 (EDT) | |||
== | === Input data used by TOMAS === | ||
#Usoskin, I. G. and Kovaltsov, G. A., ''Cosmic ray induced ionization in the atmosphere: Full modeling and practical applications'', <u>J. Geophys. Res.</u>, '''111''', doi:10.1029/2006JD007150, 2006.. | |||
#Yu, Fangqun, et al, ''Ion-mediated nucleation in the atmosphere: Key controlling parameters, implications, and look-up table'', <u>J. Geophys. Res.</u>, '''115''', D03206, doi:10.1029/2009JD012630, 2010. | |||
--[[User: | --[[User:Bmy|Bob Y.]] 17:03, 24 February 2014 (EST) | ||
Latest revision as of 21:26, 1 November 2024
This page describes the TOMAS aerosol microphysics option in GEOS-Chem. TOMAS is one of two aerosol microphysics packages being incorporated into GEOS-Chem, the other being APM.
Overview
The TwO-Moment Aerosol Sectional (TOMAS) microphysics package was developed for implementation into GEOS-Chem at Carnegie-Mellon University. Using a moving sectional and moment-based approach, TOMAS tracks two independent moments (number and mass) of the aerosol size distribution for a number of discrete size bins. It also contains codes to simulate nucleation, condensation, and coagulation processes. The aerosol species that are considered with high size resolution are sulfate, sea-salt, OC, EC, and dust. An advantage of TOMAS is the full size resolution for all chemical species and the conservation of aerosol number, the latter of which allows one to construct aerosol and CCN number budgets that will balance.
Authors and collaborators
- Peter Adams (Carnegie-Mellon U.) -- Principal Investigator
- Win Trivitayanurak (Chulalongkorn University, Thailand)
- Dan Westervelt (Princeton University, formerly Carnegie-Mellon U.)
- Jeffrey Pierce (CSU/Dalhousie U.)
- Jack Kodros (CSU)
- Salvatore Farina (Colorado State U.)
- Marguerite Marks (CMU)
--Dan W. 11:53, 27 January 2010 (EST)
TOMAS User Groups
| User Group | Personnel | Projects |
|---|---|---|
| Carnegie-Mellon University | Peter Adams Dan Westervelt |
New particle formation evaluation in GC-TOMAS Sensitivity of CCN to nucleation rates Development of number tagging and source apportionment model for GC-TOMAS |
| Colorado State | Jeffrey Pierce Sal Farina Jack Kodros |
Sensitivity of CCN to condensational growth rates TOMAS parallelization Aerosol radiative effects |
| Chulalongkorn University | Win Trivitayanurak | Source apportionment for Southeast Asia Improving size fraction information for emissions Development of WRF-GC-TOMAS |
| Add yours here |
--Bob Y. 16:35, 12 May 2014 (EDT) --Win T. 4:24, 2 Nov 2024 (GMT+7)
TOMAS-specific setup
TOMAS has its own run directories (run.Tomas) that can be downloaded from the Harvard FTP. The input.geos file will look slightly different from standard GEOS-Chem, and between versions.
Pre- v9.02: To turn on TOMAS, see the "Microphysics menu" in input.geos and make sure TOMAS is set to T.
v9.02 and later: TOMAS is enabled or disabled at compile time - the TOMAS flag in input.geos has been removed.
TOMAS is a simulation type 3 and utilizes 171-423 tracers. Each aerosol species requires 30 tracers for the 30 bin size resolution, 12 for the 12 bin, etc. Here is the (abbreviated) default setup in input.geos for TOMAS-30 in v9.02 and later (see run.Tomas directory):
Tracer # Description
1- 62 Std Geos Chem
63 H2SO4
64- 93 Number
94-123 Sulfate
124-153 Sea-salt
154-183 Hydrophilic EC
184-213 Hydrophobic EC
214-243 Hydrophilic OC
244-273 Hydrophobic OC
274-303 Mineral dust
304-333 Aerosol water
TOMAS-40 requires 423 tracers (~360 TOMAS tracers for each of the 40-bin species, and ~62 standard GEOS-Chem tracers)
--Salvatore Farina 18:48, 8 July 2013 (EDT)
Implementation notes
- The original implementation and validation of TOMAS had been done for version GEOS-Chem v8-03-01, which was released on 24 Feb 2010.
- TOMAS was completely re-integrated into GEOS-Chem v9-02, which was released on 03 Mar 2014.
- TOMAS was re-integrated into GEOS-Chem v10-01 to become compatible with HEMCO.
- Updates to TOMAS to use the SOAP/SOAS tracers were made for GEOS-Chem v11-02 (not yet released).
Update April 2013
This update was tested in the 1-month benchmark simulation v9-02k and approved on 07 Jun 2013.
Sal Farina has been working with the GEOS-Chem Support Team to inline the TOMAS aerosol microphysics code into the GeosCore directory. All TOMAS-specific sections of code are now segregated from the rest of GEOS-Chem with C-preprocessor statements such as:
#if defined( TOMAS ) # if defined( TOMAS40 ) ... Code for 40 bin TOMAS simulation (optional) goes here ... # elif defined( TOMAS12 ) ... Code for 12 bin TOMAS simulation (optional) goes here ... # elif defined( TOMAS15 ) ... Code for 15 bin TOMAS simulation (optional) goes here ... # else ... Code for 30 bin TOMAS simulation (default) goes here ... # endif #endif
TOMAS is now invoked by compiling GEOS-Chem with one of the following options:
| Command | Result |
|---|---|
| make -j4 TOMAS=yes ... | Compiles GEOS-Chem for the 30 bin (default) TOMAS simulation |
| make -j4 TOMAS12=yes ... | Compiles GEOS-Chem for the 12 bin (optional) TOMAS simulation |
| make -j4 TOMAS15=yes ... | Compiles GEOS-Chem for the 15 bin (optional) TOMAS simulation |
| make -j4 TOMAS40=yes ... | Compiles GEOS-Chem for the 40 bin (optional) TOMAS simulation |
The -j4 in the above examples tell the GNU Make utility to compile 4 files at a time. This reduces the overall compilation time.
All files in the old GeosTomas/ directory have now been deleted, as these have been rendered obsolete.
These updates are included in GEOS-Chem v9-02. These modifications will not affect the existing GEOS-Chem simulations, as all TOMAS code is not compiled into the executable unless you compile with one of the TOMAS options (described in the above table) at compile time.
--Salvatore Farina 13:49, 4 June 2013 (EDT)
--Bob Y. 16:39, 12 May 2014 (EDT)
Computational Information
GC-TOMAS v9-02 (30 sections) on 8 processors:
- One year simulation = 7-8 days wall clock time
More speedups are available using lower aerosol size resolution
--Dan W. 11:00, 07 May 2013 (EST)
GC-TOMAS v9-02 on 16 processors (glooscap)
| Simulation | Wall time / simulation year |
|---|---|
| TOMAS12 (optional) | 2.8 days |
| TOMAS15 (optional) | 3.3 days |
| TOMAS30 (default) | 6.1 days |
| TOMAS40 (optional) | 7.8 days |
--Salvatore Farina 15:51, 3 March 2014 (EST)
Microphysics Code
The aerosol microphysics code is largely contained within the file tomas_mod.f. Tomas_mod and its subroutines are modular -- they use all their own internal variables. For details, see tomas_mod.f and comments.
Nucleation
The choice of nucleation theory is selected in the header section of tomas_mod.f. The choices are currently binary homogeneous nucleation as in Vehkamaki, 2001 or ternary homogenous nucleation as in Napari et al., 2002. The ternary nucleation rate is typically scaled by a globally uniform tuning factor of 10^-4 or 10^-5. Binary nucleation (Vehkamaki et al. 2002), ion-mediated nucleation (Yu, 2008) and activation nucleation (Kulmala, 2006) are options as well.
In TOMAS-12 and TOMAS-30, nucleated particles follow the Kerminen approximation to grow to the smallest size bin. This has a tendency to overpredict the number of particles in the smallest bins of those models. See Y. H. Lee, J. R. Pierce, and P. J. Adams 2013 here for more details on the consequences of this.
Condensation
Coagulation
--Dan W. 14:08, 9 May 2011 (EST)
Validation
The following figure documents the performance of GEOS-Chem-TOMAS for predicting aerosol number (N10 = number of particles larger than 10 nm etc.) against measurements at 20 global sites. Details of observations are in
- D'Andrea, S. D., Hakkinen, S. A. K., Westervelt, D. M., Kuang, C., Levin, E. J. T., Kanawade, V. P., Leaitch, W. R., Spracklen, D. V., Riipinen, I., and Pierce, J. R.: Understanding global secondary organic aerosol amount and size-resolved condensational behavior, Atmos. Chem. Phys., 13, 11519-11534, doi:10.5194/acp-13-11519-11534, 2013.
An updated version of this figure is included as Figure 1 in Kodros and Pierce, (2017). Please note that the figure in this paper uses GEOS-Chem v10-01, which included substantial updates to emission inventories (as part of the HEMCO update). Thus, the differences in the comparisons between GEOS-Chem v8-02, v9-03, and v10-01 shown here are not necessarily due to the TOMAS code alone.
- Kodros, J. K. and Pierce, J. R.: Important global and regional differences in aerosol cloud-albedo effect estimates between simulations with and without prognostic aerosol microphysics, J. Geophys. Res. Atmos., doi:10.1002/2016JD025886, 2017
--Jeff Pierce 13:21, 4 March 2014 (MST)
--Jack Kodros 11:30, 11 June 2018 (MST)
Other features of TOMAS
Other varieties of TOMAS are suited for specific science questions, for example with nucleation studies where explicit aerosol dynamics are needed for nanometer-sized particles.
Set-up Guide
This TOMAS setup guide was written for users on ACE-NET's Glooscap cluster, but may be more generally applicable.
--Salvatore Farina 11:55, 26 July 2013 (EDT)
Size Resolution
The different TOMAS simulations (12, 15, 30, 40 size bins) have the following characteristics:
| Simulation | Size resolution |
|---|---|
| TOMAS12 | All 7 chemical species have size resolution ranging from 10 nm to 1 µm spanned by 10 logarithmically spaced (mass quadrupling) bins and two supermicron bins. Coarser resolution than TOMAS30 - Improved computation time. |
| TOMAS15 | Same as TOMAS12 with 3 additional (mass quadrupling) sub-10nm bins with a lower limit ~2nm. Analogous to TOMAS40 with improved computation time. |
| TOMAS30 | All 7 chemical species have size resolution ranging from 10 nm to 10 µm, spanned by 30 logarithmically spaced (mass doubling) bins. |
| TOMAS40 | Same as TOMAS30 with 10 additional (mass doubling) sub-10nm bins with a lower limit ~1nm. |
--Salvatore Farina 12:51, 4 June 2013 (EDT)
Nesting and grid size
As of v10.01, TOMAS has been implemented and tested on the 4x5, 2x2.5, 0.5x0.667 (North America and Asia), and 0.25x0.3125 (Asian) domains. To the best of our knowledge, these grids should continue working in upcoming releases of GEOS-Chem (including the grid-independent GEOS-Chem).
Sample code
To assist new users, sample processing code (in Python) is available for GEOS-Chem-TOMAS output. Specifically, this code reads in GEOS-Chem-TOMAS output and demonstrates some common calculations (such as calculating size distributions, bin diameters, CCN, etc.). The primary focus of this code is to provide a few examples rather than a complete, efficient package. Further, this code was written based on GEOS-Chem-TOMAS v10.01, which outputs monthly tracers in bpch format. We also provide sample IDL scripts to convert bpch files to netCDF using GAMAP routines. Future versions of GEOS-Chem will output netCDF directly, and so these IDL routines will become obsolete.
Git repository for sample TOMAS code: https://bitbucket.org/teampierce/sample_processing/src/master/
--Jack Kodros 1:12, 11 June 2018 (MST)
AOD, CCN post-processing code
Codes available for calculating aerosol optical depth using TOMAS predicted aerosol composition and size and Mie Theory. Also CCN concentrations calculated from TOMAS size-resolved composition and Kohler theory. Developed by Yunha Lee and Jeffrey Pierce, adapted for GEOS-Chem output by Jeffrey Pierce.
--Dan W. 2:00, 9 May 2011 (EST)
Biomass burning subgrid coagulation switch
This update was included in GEOS-Chem 12.2.1, which was released on 28 Feb 2019.
Emily Ramnarine created code that allows the "emitted" size distribution in the model be a function of a number of properties that include the mean emissions rate per fire in the grid box. In order to do this, Emily needed to (1) make changes to carbon_mod.F, (2) add a line or few to HEMCO_config.rc, and (3) modify the FINN input files to include the number of fires. This only affects TOMAS simulations. Emily wrote:
- This parameterization, based on Sakamoto et al (2016), estimates the amount of near-source, sub-grid scale coagulation happening in a biomass burning plume. Can be turned on or off. When on, the default assumption is that each smoke plume is completely seperate from the others (i.e. there is no overlap of the plumes). There is also an option for all smoke plumes in a grid box to overlap completely. When being used, this parameterization changes the median diameter and modal width of biomass burning emissions to account for coagulation.
Reference Ramnarine, E., Kodros, J. K., Hodshire, A. L., Lonsdale, C. R., Alvarado, M. J., and Pierce, J. R., Effects of Near-Source Coagulation of Biomass Burning Aerosols on Global Predictions of Aerosol Size Distributions and Implications for Aerosol Radiative Effects, Atmos. Chem. Phys. Discuss., https://doi.org/10.5194/acp-2018-1084, in review, 2018.
--Melissa Sulprizio (talk) 15:07, 22 February 2019 (UTC)
--Bob Yantosca (talk) 18:54, 28 February 2019 (UTC)
References
In this section we provide references relevant to TOMAS aerosl microphysics simulations.
Studies using TOMAS simulations
- Nucleation in GEOS-Chem: Westervelt, D. M., Pierce, J. R., Riipinen, I., Trivitayanurak, W., Hamed, A., Kulmala, M., Laaksonen, A., Decesari, S., and Adams, P. J.: Formation and growth of nucleated particles into cloud condensation nuclei: model-measurement comparison, Atmos. Chem. Phys. Discuss., 13, 8333-8386, doi:10.5194/acpd-13-8333-2013, 2013. LINK
- TOMAS implementation in GEOS-Chem: Trivitayanurak, W., Adams, P. J., Spracklen, D. V. and Carslaw, K. S.: Tropospheric aerosol microphysics simulation with assimilated meteorology: model description and intermodel comparison, Atmos. Chem. Phys., 8(12), 3149-3168, 2008.
- TOMAS initial paper, sulfate only: Adams, P. J. and Seinfeld, J. H.: redicting global aerosol size distributions in general circulation models, J. Geophys. Res.-Atmos., 107(D19), -, doi:Artn 4370 Doi 10.1029/2001jd001010, 2002.
- TOMAS with sea-salt: Pierce, J.R., and Adams P.J., Global evaluation of CCN formation by direct emission of sea salt and growth of ultrafine sea salt, J. Geophys. Res.-Atmos., 111 (D6), doi:10.1029/2005JD006186, 2006.
- TOMAS with carbonaceous aerosol: Pierce, J. R., Chen, K. and Adams, P. J.: Contribution of primary carbonaceous aerosol to cloud condensation nuclei: processes and uncertainties evaluated with a global aerosol microphysics model, Atmos. Chem. Phys., 7(20), 5447-5466, doi:10.5194/acp-7-5447-2007, 2007.; Trivitayanurak, W. and Adams, P. J.: Does the POA–SOA split matter for global CCN formation?, Atmos. Chem. Phys., 14, 995–1010, doi:10.5194/acp-14-995-2014, 2014.
- TOMAS with dust: Lee, Y.H., K. Chen, and P.J. Adams, 2009: Development of a global model of mineral dust aerosol microphysics. Atmos. Chem. Phys., 8, 2441-2558, doi:10.5194/acp-9-2441-2009.
- TOMAS with SOA: D'Andrea, S. D., Hakkinen, S. A. K., Westervelt, D. M., Kuang, C., Levin, E. J. T., Kanawade, V. P., Leaitch, W. R., Spracklen, D. V., Riipinen, I., and Pierce, J. R.: Understanding global secondary organic aerosol amount and size-resolved condensational behavior, Atmos. Chem. Phys., 13, 11519-11534, doi:10.5194/acp-13-11519-11534, 2013.
- TOMAS with offline DRE/AIE: Kodros, J. K., Cucinotta, R., Ridley, D. A., Wiedinmyer, C. and Pierce, J. R.: The aerosol radiative effects of uncontrolled combustion of domestic waste, Atmos. Chem. Phys., 16(11), 6771-6784, doi:10.5194/acp-16- 6771-2016, 2016
- TOMAS compared to GEOS-Chem standard: Kodros, J. K. and Pierce, J. R.: Important global and regional differences in aerosol cloud-albedo effect estimates between simulations with and without prognostic aerosol microphysics, J. Geophys. Res. Atmos., doi:10.1002/2016JD025886, 2017.
--Bob Y. 09:53, 2 June 2014 (EDT), Win T. 17:18, 28 June 2021 (EDT)
Input data used by TOMAS
- Usoskin, I. G. and Kovaltsov, G. A., Cosmic ray induced ionization in the atmosphere: Full modeling and practical applications, J. Geophys. Res., 111, doi:10.1029/2006JD007150, 2006..
- Yu, Fangqun, et al, Ion-mediated nucleation in the atmosphere: Key controlling parameters, implications, and look-up table, J. Geophys. Res., 115, D03206, doi:10.1029/2009JD012630, 2010.
--Bob Y. 17:03, 24 February 2014 (EST)